Abstract
All xenografts from pigs impose infection risk by porcine endogenous retrovirus (PERV). The purpose of this study was to investigate the env constructs with the comparison of the ratio of the competent form to the defective one of env in subtypes, PERV-A, PERV-B and PERV-C in different pig breeds. The results of PCR amplification of env represented that all env subtypes had more than two defective forms which cannot bind to host cells due to the absence of binding regions of env in miniature pigs, SNU and PWG, and farm pig breeds, Duroc, Yorkshire and Landrace. In addition, comparing the full sequences with the defective ones in three subtypes demonstrated that the present percentages of env sequences in defective PERV-A, PERV-B and PERV-C were approximately 50%, 38∼45% and 4∼11%, respectively, in SNU and PWG pigs whereas PERV-A and PERV-B occupied around 40 to 60% but PERV-C was not detected in farm pigs. Quantitative real-time PCR assays with primers and probes targeted to proline-rich region (PRR) of each env subtype were done to measure the copy numbers of each env subtype. When the reference was set with copy number of PERV-A, the ratio of those of PERV-B and PERV-C to the reference were 1.5 to 6.0 folds high in SNU and PWG pigs while 1.0 or less in farm pigs. These contradictory results of PERV-C constructs and copy numbers in SNU pigs suggests that many truncated or short defective sequences of PERV-C might be present in them.
Go to : 
REFERENCES
1). Martignat L, Saï P, Jestin A. Detection of porcine endogenous retrovirus: possible involvement in pig islet xenotransplantation. Diabetes Metab. 1998; 24:434–41.
2). Denner J, Tönjes RR. Infection barriers to successful xenotransplantation focusing on porcine endogenous retroviruses. Clin Microbiol Rev. 2012; 25:318–43.

3). Schmidt P, Andersson G, Blomberg J, Malmsten A, Korsgren O. Possible transmission of zoonoses in xenotransplantation: porcine endogenous retroviruses (PERVs) from an immunological point of view. Acta Vet Scand Suppl. 2004; 99:27–34.

4). Frühauf JH, Mertsching H, Giri S, Frühauf NR, Bader A. Porcine endogenous retrovirus released by a bioartificial liver infects primary human cells. Liver Int. 2009; 29:1553–61.

5). Martin U, Kiessig V, Blusch JH, Haverich A, von der Helm K, Herden T, et al. Expression of pig endogenous retrovirus by primary porcine endothelial cells and infection of human cells. Lancet. 1998; 352:692–4.

6). Ritzhaupt A, Van Der Laan LJ, Salomon DR, Wilson CA. Porcine endogenous retrovirus infects but does not replicate in nonhuman primate primary cells and cell lines. J Virol. 2002; 76:11312–20.

7). Kim JH, Jung ES, Park CG, Kim SJ, Hwang ES. No Evidence of the Productive Replication of Porcine Endogenous Retrovirus (PERV) from SNU Miniature Pigs in Human Cell Line. Infect Chemother. 2010; 42:175–80.

8). Switzer WM, Michler RE, Shanmugam V, Matthews A, Hussain AI, Wright A, et al. Lack of cross-species transmission of porcine endogenous retrovirus infection to nonhuman primate recipients of porcine cells, tissues, or organs. Transplantation. 2001; 71:959–65.
9). Walles T, Lichtenberg A, Puschmann C, Leyh R, Wilhelmi M, Kallenbach K, et al. In vivo model for cross-species porcine endogenous retrovirus transmission using tissue engineered pulmonary arteries. Eur J Cardiothorac Surg. 2003; 24:358–63.
10). Moalic Y, Blanchard Y, Félix H, Jestin A. Porcine endogenous retrovirus integration sites in the human genome: features in common with those of murine leukemia virus. J Virol. 2006; 80:10980–8.

11). Takeuchi Y, Patience C, Magre S, Weiss RA, Banerjee PT, Le Tissier P, et al. Host range and interference studies of three classes of pig endogenous retrovirus. J Virol. 1998; 72:9986–91.

12). Popp SK, Mann DA, Milburn PJ, Gibbs AJ, McCullagh PJ, Wilson JD, et al. Transient transmission of porcine endogenous retrovirus to fetal lambs after pig islet tissue xenotransplantation. Immunol Cell Biol. 2007; 85:238–48.

13). Denner J. Recombinant porcine endogenous retroviruses (PERV-A/C): a new risk for xenotransplantation? Arch Virol. 2008; 153:1421–6.

14). Pinter A, Fleissner E. The presence of disulfide-linked gp70-p15 (E) complexes in AKR murine leukemia virus. Virology. 1977; 83:417–22.
15). Argaw T, Figueroa M, Salomon DR, Wilson CA. Identification of residues outside of the receptor binding domain that influence the infectivity and tropism of porcine endogenous retrovirus. J Virol. 2008; 82:7483–91.

16). Argaw T, Wilson CA. Detailed Mapping of Determinants within the Porcine Endogenous Retrovirus Envelope Surface Unit Identifies Critical Residues for Human Cell Infection within the Proline-Rich Region. J Virol. 2012; 86:9096–104.

17). Mazurek U, Kimsa MC, Strzalka-Mrozik B, Kimsa MW, Adamska J, Lipinski D, et al. Quantitative analysis of porcine endogenous retroviruses in different organs of transgenic pigs generated for xenotransplantation. Curr Microbiol. 2013; 67:505–14.

18). Kaulitz D, Mihica D, Adlhoch C, Semaan M, Denner J. Improved pig donor screening including newly identified variants of porcine endogenous retrovirus-C (PERV-C). Arch Virol. 2013; 158:341–8.

19). Mang R, Maas J, Chen X, Goudsmit J, van Der Kuyl AC. Identification of a novel type C porcine endogenous retrovirus: evidence that copy number of endogenous retroviruses increases during host inbreeding. J Gen Virol. 2001; 82:1829–34.

20). Quereda JJ, Herrero-Medrano JM, Abellaneda JM, García-Nicolás O, Martínez-Alarcón L, Pallarés FJ, et al. Porcine endogenous retrovirus copy number in different pig breeds is not related to genetic diversity. Zoonoses Public Health. 2012; 59:401–7.

21). Schuurman HJ. The International Xenotransplantation Association consensus statement on conditions for undertaking clinical trials of porcine islet products in type 1 diabetes–chapter 2: Source pigs. Xenotransplantation. 2009; 16:215–22.
22). Büscher K, Hahn S, Hofmann M, Trefzer U, Ozel M, Sterry W, et al. Expression of the human endogenous retrovirus-K transmembrane envelope, Rec and Np9 proteins in melanomas and melanoma cell lines. Melanoma Res. 2006; 16:223–34.

23). Chen T, Meng Z, Gan Y, Wang X, Xu F, Gu Y, et al. The viral oncogene Np9 acts as a critical molecular switch for co-activating beta-catenin, ERK, Akt and Notch1 and promoting the growth of human leukemia stem/progenitor cells. Leukemia. 2013; 27:1469–78.
24). Gross H, Barth S, Pfuhl T, Willnecker V, Spurk A, Gurtsevitch V, et al. The NP9 protein encoded by the human endogenous retrovirus HERV-K (HML-2) negatively regulates gene activation of the Epstein-Barr virus nuclear antigen 2 (EBNA2). Int J Cancer. 2011; 129:1105–15.
25). Mayer J, Ehlhardt S, Seifert M, Sauter M, Müller-Lantzsch N, Mehraein Y, et al. Human endogenous retrovirus HERV-K (HML-2) proviruses with Rec protein coding capacity and transcriptional activity. Virology. 2004; 322:190–8.
Go to : 
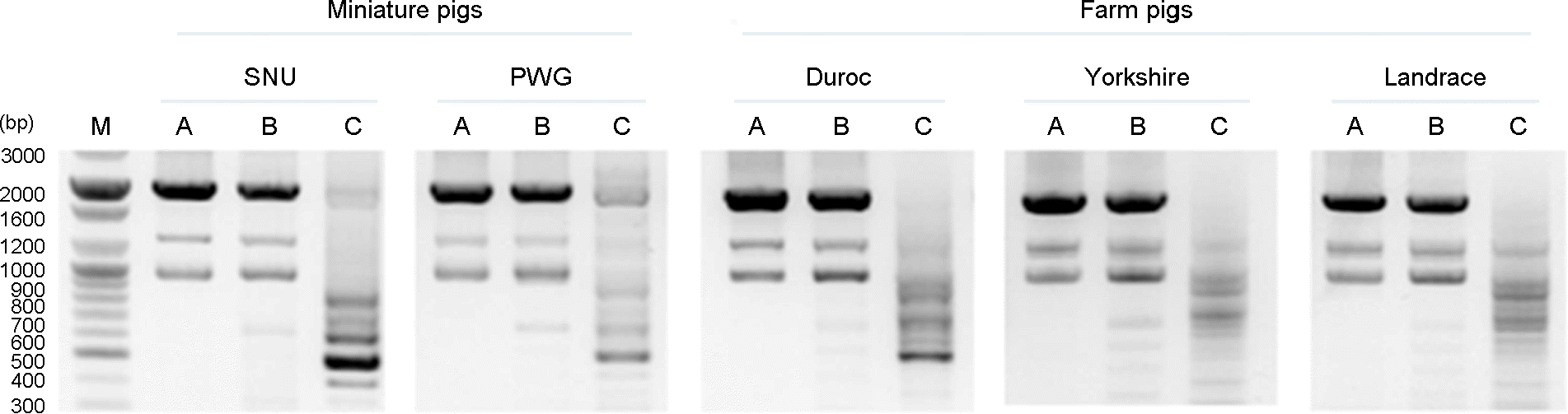 | Figure 1.Amplified profile of env subtypes in miniature pigs (SNU and PWG) and farm pig breeds (Duroc, Yorkshire and Landrace). Gene amplification was done by PCR with the primers described in Table 1. All the experiments revealed the similar amplification patterns and the representative pictures were shown. (M) denotes molecular marker; (A), (B) and (C) denote PERV subtypes A, B and C, respectively. |
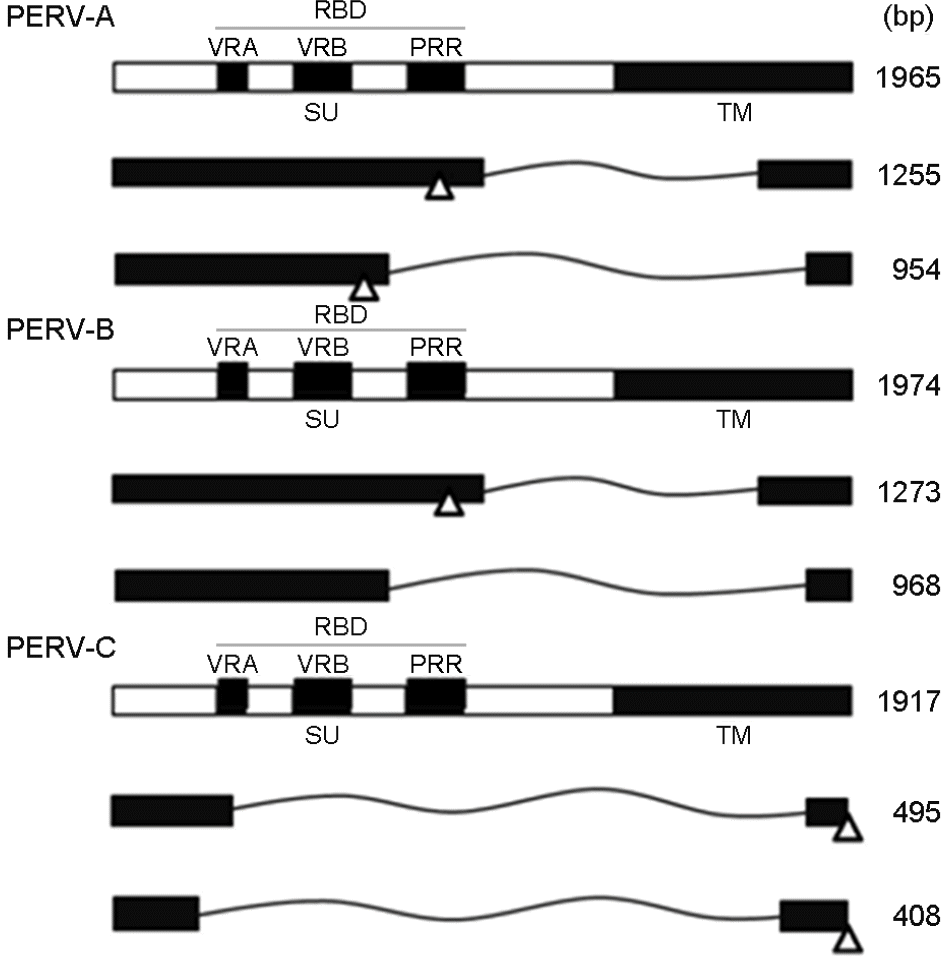 | Figure 2.Schematic diagrams of constructs of PERV A, B and C env gene derived from SNU miniature pig. SU, surface; TM, transmembrane; RBD, receptor binding domain; VRA, variable region a; VRB, variable region b; PRR, proline-rich region; open triangle: stop codon. |
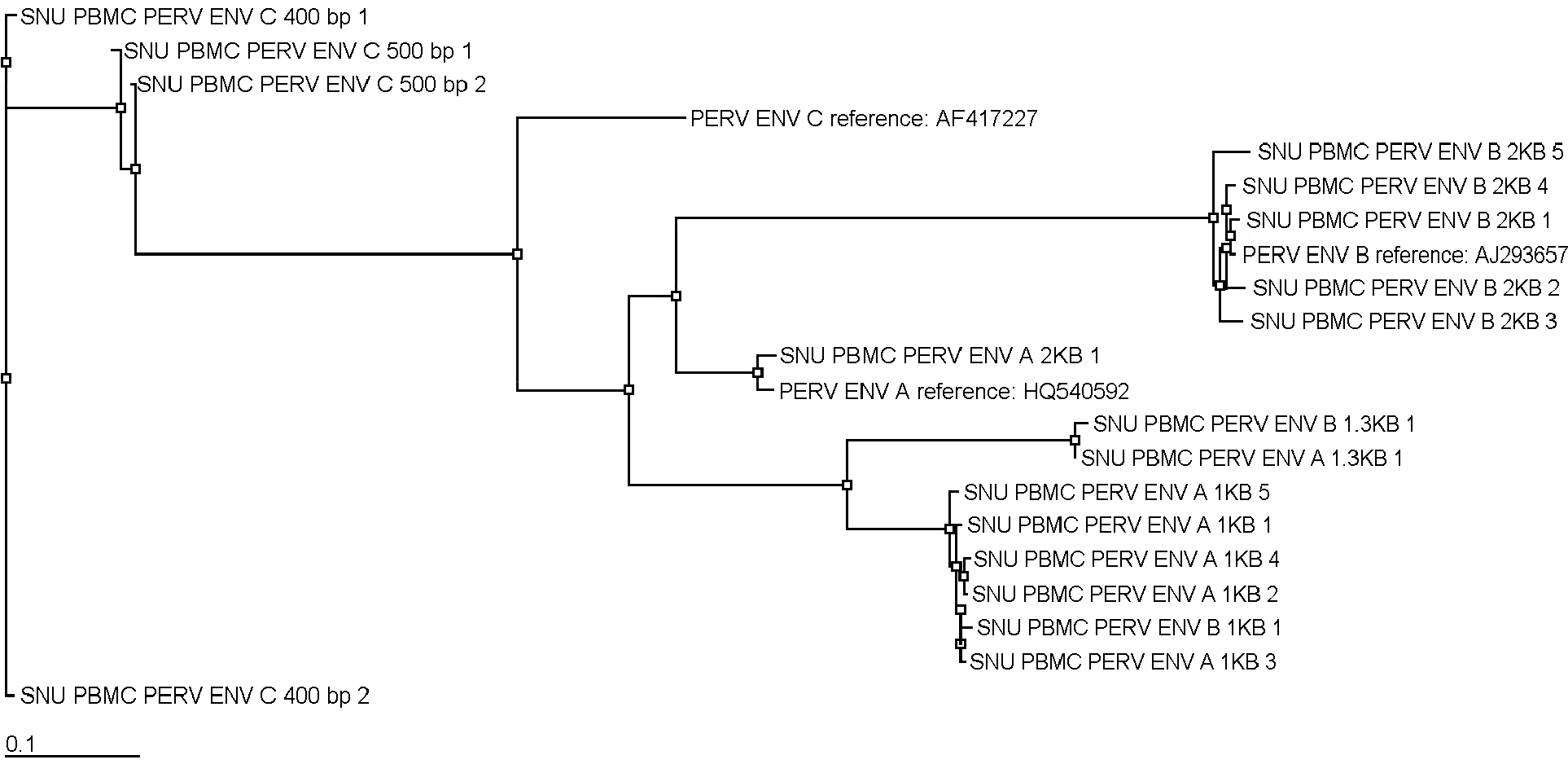 | Figure 3.Pylogenetic relationship tree using neighbor-joining methods on the nucleotide sequences of complete and defective PERV env derived from SNU pigs. |
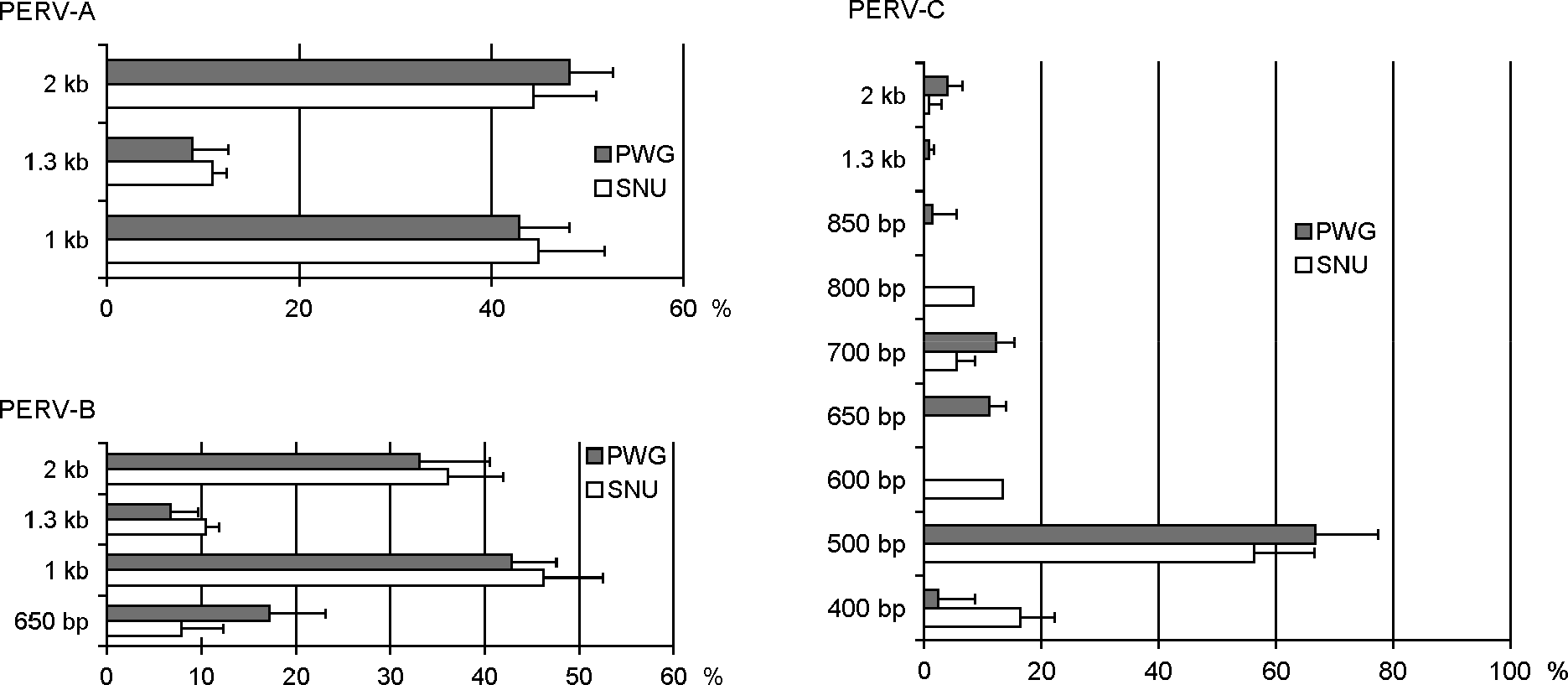 | Figure 4.The proportion of the full length sequences and defective ones of env in 18 SNU and 11 PWG miniature pigs. The relative amount of each construct was measured by the densitometry of amplified PCR products on agarose gel. Results were shown with mean + standard deviation. |
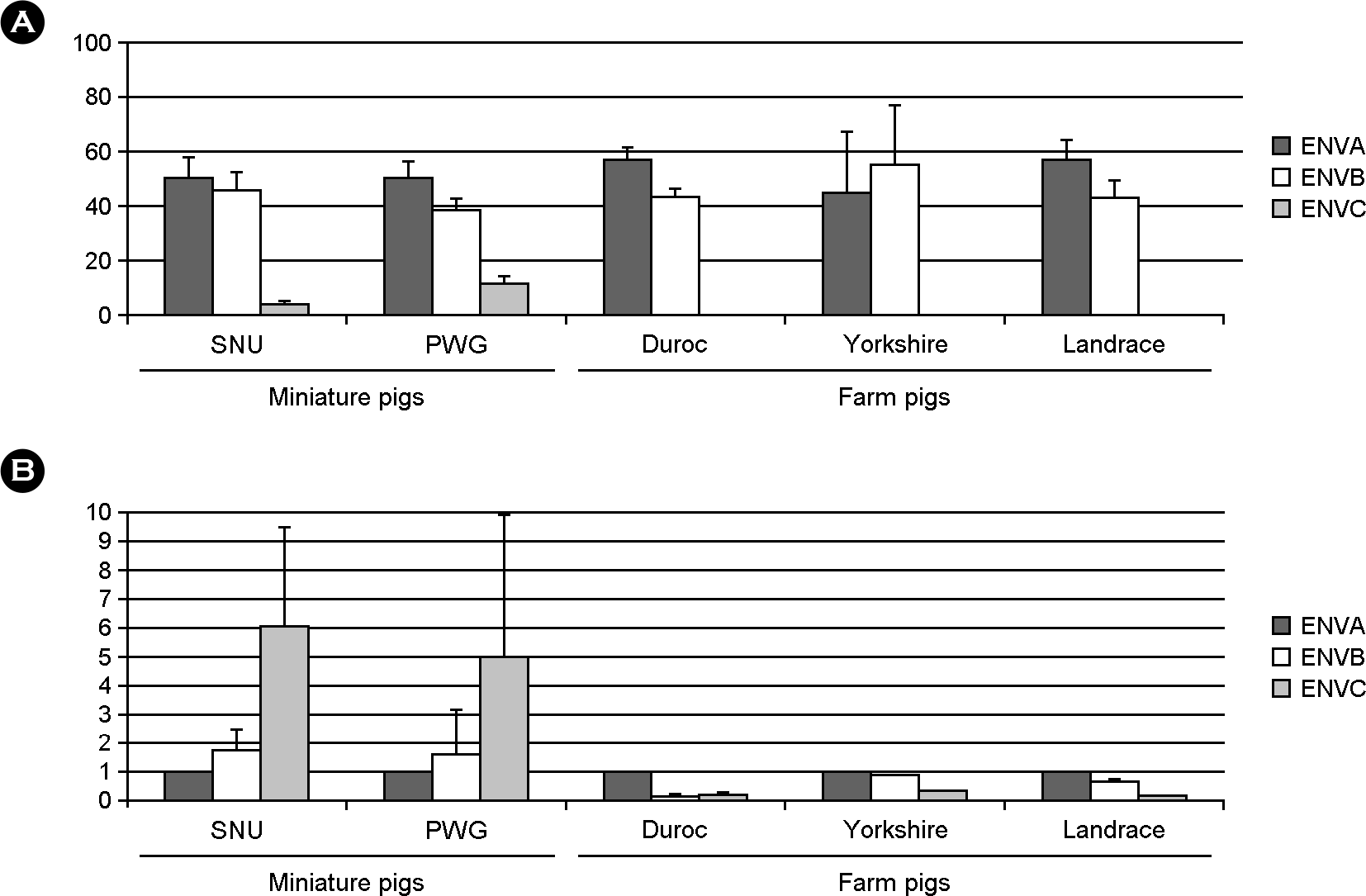 | Figure 6.The proportion of the env A, B and C subtypes in various pig breeds. (A) the percentages of each env subtype by densitometry of PCR product on the agarose gel. (B) the ratio of copy numbers of PERV-B and PERV-C to that of PERV-A. Results were shown in mean + standard deviation in 18 SNU, 11 PWG, 3 Duroc, 3 Yorkshire, and 3 Landrace pigs. |
Table 1.
PERV env specific primers and probes used in this study.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download