Abstract
To investigate the possibility of transmission of CTX-M-producing Escherichia coli isolates among humans and animals, we compared CTX-M-producing E. coli isolates showing the same genotype from humans and dogs in Korea. Sixteen CTX-M-producing E. coli isolates from animals were selected and their genotypes were identified using MLST. Among clinical CTX-M-producing E. coli isolates from humans, which have been identified in previous studies, 12 isolates showing the same STs with those of E. coli isolates from animals were selected. For these 28 CTX-M-producing E. coli isolates, identification of blaCTX-M genes and their genetic environments, antimicrobial susceptibility testing, extended MLST, and PFGE were performed. Some CTX-M-producing E. coli isolates from humans showed the same genotypes, such as ST10, ST38, ST58, and ST95, but different CTX-M enzymes and PFGE patterns. Thus, it can be concluded that dissemination of ESBL-producing E. coli isolates between humans and animals is rare so far.
Go to : 
REFERENCES
1). Hammerum AM, Heuer OE. Human health hazards from antimicrobial-resistant Escherichia coli of animal origin. Clin Infect Dis. 2009; 48:916–21.
2). Cantón R, Coque TM. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol. 2006; 9:466–75.
3). Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 2011; 66:1–14.
4). Matsumoto Y, Ikeda F, Kamimura T, Yokota Y, Mine Y. Novel plasmid-mediated beta-lactamase from Escherichia coli that inactivates oxyimino-cephalosporins. Antimicrob Agents Chemother. 1998; 32:1243–6.
5). Ewers C, Grobbel M, Stamm I, Kopp PA, Diehl I, Semmler T, et al. Emergence of human pandemic O25: H4-ST131 CTX-M-15 extended-spectrum-β-lactamase-producing Escherichia coli among companion animals. J Antimicrob Chemother. 2010; 65:651–60.
6). Baraniak A, Fiett J, Sulikowska A, Hryniewicz W, Gniadkowski M. Countrywide spread of CTX-M-3 extended-spectrum β-lactamase-producing microorganisms of the family Enterobacteriaceae in Poland. Antimicrob Agents Chemother. 2002; 46:151–9.
7). Ho PL, Lo WU, Yeung MK, Li Z, Chan J, Chow KH, et al. Dissemination of pHK01-like incompatibility group IncFII plasmids encoding CTX-M-14 in Escherichia coli from human and animal sources. Vet Microbiol. 2012; 158:172–9.
8). Guenther S, Ewers C, Wieler LH. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front Microbiol. 2011; 2:246.

9). Tamang MD, Nam HM, Jang GC, Kim SR, Chae MH, Jung SC, et al. Molecular characterization of extended-spectrum-β-lactamase-producing and plasmid-mediated AmpC β-lactamase-producing Escherichia coli isolated from stray dogs in South Korea. Antimicrob Agents Chemother. 2012; 56:2705–12.
10). Shin JY, Kim DH, Ko KS. Comparison of CTX-M-14- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae isolates from patients with bacteremia. J Infect. 2011; 63:39–47.
11). Kim J, Lim YM, Jeong YS, Seol SY. Occurrence of CTX-M-3, CTX-M-15, CTXM-14, and CTX-M-9 extended-spectrum-β-lactamases in Enterobacteriaceae clinical isolates in Korea. Antimicrob Agents Chemother. 2005; 49:1572–5.
12). Ruppé E, Hem S, Lath S, Gautier V, Ariey F, Sarthou JL, et al. CTX-M β-lactamases in Escherichia coli from community-acquired urinary tract infections, Cambodia. Emerg Infect Dis. 2009; 15:741–8.
13). Kim JM, Lim YM. Prevalence of CTX-M extended-spectrum beta-lactamases in clinical isolates of Enterobacteriaceae in Korea. J Bacteriol Virol. 2004; 34:303–10.
14). Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing; approved guideline. CLSI Document M100-S21. Clinical and Laboratory Standards Institute;Wayne, PA: 2011.
15). Choi MJ, Ko KS. Persister cells: Survival strategies under antimicrobiotic stress. J Bacteriol Virol. 2013; 43:73–6.

16). Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005; 43:4178–82.
17). Walk ST, Alm EW, Gordon DM, Ram JL, Toranzos GA, Tiedje JM, et al. Cryptic lineages of the Genus Escherichia. Appl Environ Microbial. 2009; 75:6534–44.
18). Ko KS, Lee JY, Baek JY, Suh JY, Lee MY, Choi JY, et al. Predominance of an ST11 extended-spectrum β-lactamase-producing Klebsiella pneumoniae clone causing bacteraemia and urinary tract infections in Korea. J Med Microbiol. 2010; 59:822–8.
19). Dhanji H, Patel R, Wall R, Doumith M, Patel B, Hope R, et al. Variation in the genetic environments of bla(CTX-M-15) in Escherichia coli from the faeces of travellers returning to the United Kingdom. J Antimicrob Chemother. 2011; 66:1005–12.
20). Eckert C, Gautier V, Arlet G. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J Antimicrob Chemother. 2006; 57:14–23.
21). Shakil S, Khan AU. Interaction of CTX-M-15 enzyme with cefotaxime: a molecular modeling and docking study. Bioinformation. 2010; 4:468–72.
22). Xiong ZZ, Zhu DM, Wang F, Zhang YY. CTX-M-14, CTX-M-24 and resistance in Escherichia coli and Klebsiella pneumoniae clinical isolates. Chin Med J. 2006; 119:160–4.
23). Bonnedahl J, Drobni M, Gauthier-Clerc M, Hernandez J, Granholm S, Kayser Y, et al. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the South of France. PloS One. 2009; 4:e5958.
24). Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, et al. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, the Netherlands. Emerg Infect Dis. 2011; 17:1216–22.
25). Stokes MO, Cottell JL, Piddock LJ, Wu G, Wootton M, Mevius DJ, et al. Detection and characterization of pCT-like plasmid vectors for blaCTX-M-14 in Escherichia coli isolates from humans, turkeys and cattle in England and Wales. J Antimicrob Chemother. 2012; 67:1639–44.
26). So JH, Kim J, Bae IK, Jeong SH, Kim SH, Lim SK, et al. Dissemination of multidrug-resistant Escherichia coli in Korean veterinary hospitals. Diagn Microbiol Infect Dis. 2012; 73:195–9.
27). Shin J, Choi MJ, Ko KS. Replicon sequence typing of IncF plasmids and the genetic environments of blaCTX-M-15 indicate multiple acquisitions of blaCTX-M-15 in Escherichia coli and Klebsiella pneumoniae isolates from South Korea. J Antimicrob Chemother. 2012; 67:1853–7.
Go to : 
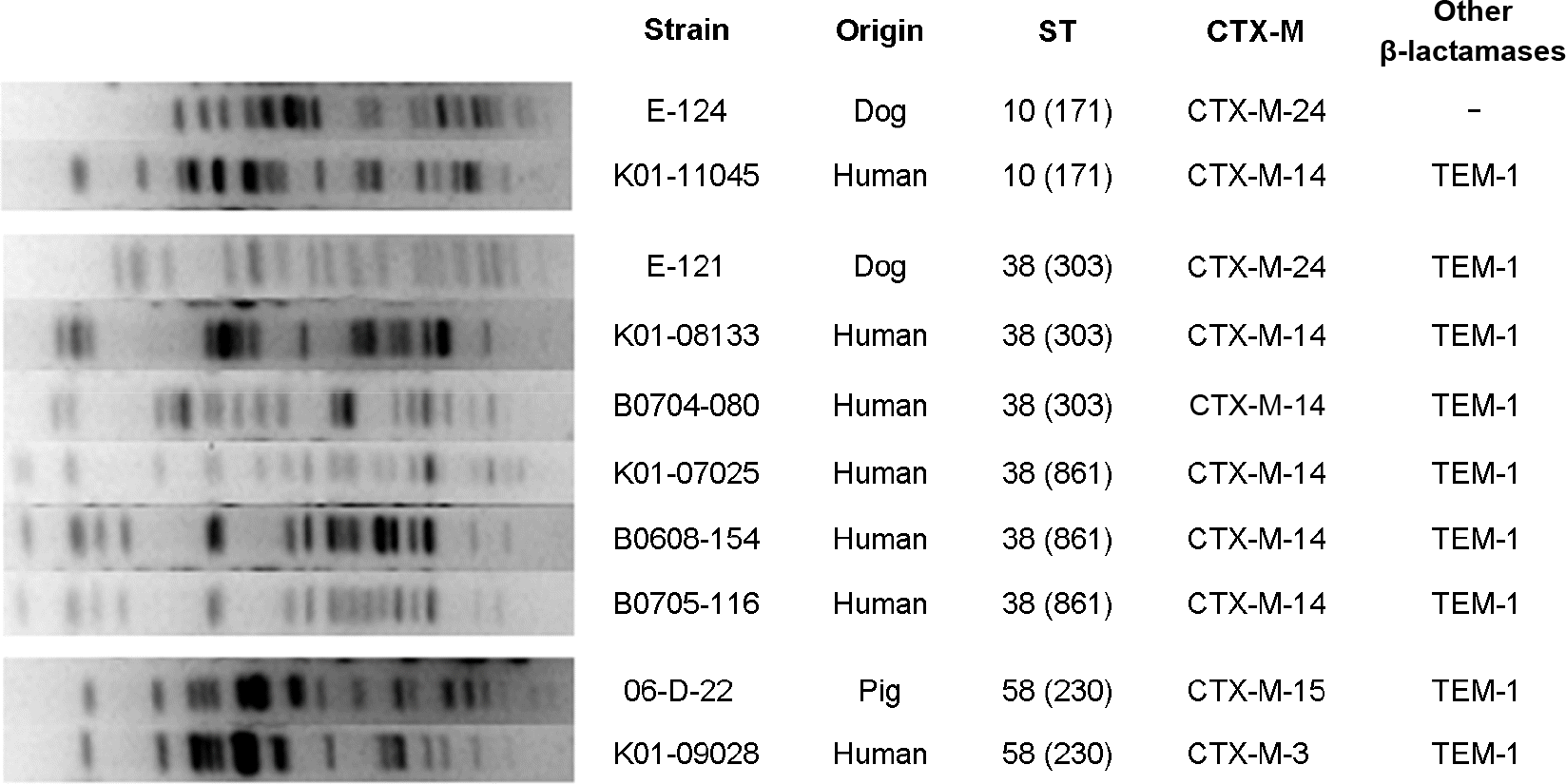 | Figure 1.Comparison of PFGE patterns of ST10, ST38, and ST58 CTX-M-producing E. coli isolates from humans and animals. |
Table 1.
ESBL types, STs, and antimicrobial resistant profiles in ESBL-producing E. coli isolates from animals.
| Strains | Origin | CTX-M | Other β-lactamases detected | ST |
Susceptibility to other antibioticsa |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK | AMP | AZT | CFX | CFZ | CIP | GEN | IMI | PIP/TZ | TMP/SMX | |||||
| CTX-M-1 group | ||||||||||||||
| E-6 | Dog | CTX-M-3 | TEM-1 | 95 | S | R | S | R | S | S | S | S | R | R |
| 06-D-22 | Pig | CTX-M-15 | TEM-1 | 58 | S | R | R | R | R | S | S | S | R | R |
| E-33 | Dog | CTX-M-55 | − | 125 | S | R | R | R | R | S | S | S | R | R |
| CTX-M-9 group | ||||||||||||||
| E-120 | Dog | CTX-M-14 | − | 457 | S | R | R | R | R | R | R | S | R | R |
| E-123 | Dog | CTX-M-14 | − | 457 | S | R | R | R | S | S | S | S | R | S |
| E-126 | Dog | CTX-M-14 | − | 2,541 | S | R | R | R | S | S | S | S | R | R |
| E-128 | Dog | CTX-M-14 | TEM-1 | 359 | R | R | R | R | S | R | R | S | R | R |
| E-129 | Dog | CTX-M-14 | TEM-1 | 93 | S | R | R | R | S | R | S | S | R | R |
| 04-D-02 | Dog | CTX-M-14 | − | 327 | S | R | R | R | S | S | S | S | R | S |
| 04-D-26 | Cattle | CTX-M-14 | TEM-1 | 2,197 | S | R | R | R | S | R | R | S | R | R |
| 05-D-37 | Cattle | CTX-M-14 | TEM-1 | 2,930 | R | R | R | R | I | R | R | S | R | R |
| E-102 | Dog | CTX-M-24 | − | 642 | S | R | R | R | S | S | S | S | R | R |
| E-121 | Dog | CTX-M-24 | TEM-1 | 38 | S | R | R | R | R | R | R | S | R | R |
| E-124 | Dog | CTX-M-24 | − | 10 | S | R | R | R | S | R | S | S | R | S |
| E-122 | Dog | CTX-M-27 | − | 457 | S | R | R | R | R | R | R | S | R | R |
| E-127 | Dog | CTX-M-65 | − | 327 | I | R | R | R | S | S | R | S | R | S |
Table 2.
ESBL types, STs, and antimicrobial resistant profiles in ESBL-producing E. coli isolates from human.
| Strains | CTX-M | Other β-lactamases detected | ST |
Susceptibility to other antibioticsa |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK | AMP | AZT | CFX | CFZ | CIP | GEN | IMI | PIP/TZ | TMP/SMX | ||||
| CTX-M-1 group | |||||||||||||
| K01-09028 | CTX-M-3 | TEM-1 | 58 | R | R | R | R | R | R | S | S | R | R |
| B0707-025 | CTX-M-15 | TEM-1 | 38 | S | R | R | R | R | R | R | S | R | R |
| CTX-M-9 group | |||||||||||||
| K01-07025 | CTX-M-14 | TEM-1 | 38 | S | R | R | R | R | R | I | S | R | R |
| K01-08133 | CTX-M-14 | TEM-1 | 38 | R | R | R | R | R | R | R | S | R | R |
| B0608-154 | CTX-M-14 | TEM-1 | 38 | S | R | R | R | I | R | R | S | S | S |
| B0704-080 | CTX-M-14 | TEM-1 | 38 | S | R | I | R | S | S | S | S | S | R |
| B0705-116 | CTX-M-14 | TEM-1 | 38 | S | R | R | R | S | S | R | S | S | R |
| K01-09061 | CTX-M-14 | TEM-1 | 95 | S | R | R | R | R | I | R | S | R | R |
| K01-12012 | CTX-M-14 | TEM-1 | 95 | I | R | R | R | R | R | S | S | R | S |
| B0610-104 | CTX-M-14 | TEM-1 | 95 | R | R | R | S | S | S | R | S | S | R |
| B0702-129 | CTX-M-14 | TEM-1 | 95 | S | R | R | R | S | S | S | S | S | S |
| K01-11045 | CTX-M-14 | TEM-1 | 10 | I | R | R | R | R | R | R | S | R | R |
Table 3.
Genotypic characterization of blaCTX-M surrounding DNA




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download