Abstract
Hepatitis B virus (HBV) and hepatitis C virus (HCV) infections are among the most common causes of chronic liver disease worldwide. The host immune pressure against hepatitis viruses during the chronic infection has led to mutations in their coding genes, which could play a pivotal role in the clinical outcomes of chronic patients. Our recent molecular epidemiologic studies regarding the HBV precore/core (preC/C) regions and HCV nonstructural 5B (NS5B) protein suggest the presence of distinct CD4 T cell immune pressure against HBV and HCV in Korean chronic patients. However, induced HBV and HCV mutations seem to exert an opposite effect on Korean chronic hepatitis B (CHB) and chronic hepatitis C (CHC) patients, respectively. On the basis of two of our recent papers, we focused in this review on the relationships between the mutation patterns of HBV preC/C and HCV NS5B, which were presumed to be caused by distinct CD4 T cell pressure in the Korean population and their effect on the clinical outcomes and liver disease progression of CHB and CHC patients.
Go to : 
REFERENCES
2). KCDC. Report of National Health and Nutrition Survay. 2010.
4). Kidd-Ljunggren K, Miyakawa Y, Kidd AH. Genetic variability in hepatitis B viruses. J Gen Virol. 2002; 83:1267–80.

6). Kim H, Jee YM, Song BC, Hyun JW, Mun HS, Kim HJ, et al. Analysis of hepatitis B virus quasispecies distribution in a Korean chronic patient based on the full genome sequences. J Med Virol. 2007; 79:212–9.

7). Bronowicki JP, Ouzan D, Asselah T, Desmorat H, Zarski JP, Foucher J, et al. Effect of Ribavirin in Genotype 1 Patients With Hepatitis C Responding to Pegylated Interferon Alfa-2a Plus Ribavirin. Gastroenterology. 2006; 131:1040–8.

8). Kwo PY, Lawitz EJ, McCone J, Schiff ER, Vierling JM, Pound D, et al. Efficacy of boceprevir, an NS3 protease inhibitor, in combination with peginterferon alfa-2b and ribavirin in treatment-naive patients with genotype 1 hepatitis C infection (SPRINT-1): an open-label, randomised, multicentre phase 2 trial. Lancet. 2010; 376:705–16.

9). Lee DS, Sung YC, Whang YS. Distribution of HCV genotypes among blood donors, patients with chronic liver disease, hepatocellular carcinoma, and patients on maintenance hemodialysis in Korea. J Med Virol. 1996; 49:55–60.

10). Kim BJ, Song BC. Distribution of hepatitis B virus genotypes according to the clinical outcomes in patients with chronic hepatitis B virus infection in Jeju island. Korean J Gastroenterol. 2003; 42:496–501.
11). Kim DW, Lee SA, Hwang ES, Kook YH, Kim BJ. Naturally occurring precore/core region mutations of hepatitis B virus genotype C related to hepatocellular carcinoma. PLoS One. 2012; 7:e47372.

12). Kim H, Jee Y, Mun HS, Park JH, Yoon JH, Kim YJ, et al. Characterization of two hepatitis B virus populations in a single Korean hepatocellular carcinoma patient with an HBeAg-negative serostatus: a novel X-Gene-deleted strain with inverted duplication sequences of upstream enhancer site II. Intervirology. 2007; 50:273–80.

13). Kim H, Jee Y, Mun HS, Song BC, Park JH, Hyun JW, et al. Comparison of full genome sequences between two hepatitis B virus strains with or without preC mutation (A1896) from a single Korean hepatocellular carcinoma patient. J Microbiol Biotechnol. 2007; 17:701–4.
14). Kim H, Jee YM, Song BC, Shin JW, Yang SH, Mun HS, et al. Molecular epidemiology of hepatitis B virus (HBV) genotypes and serotypes in patients with chronic HBV infection in Korea. Intervirology. 2007; 50:52–7.

15). Kim H, Lee SA, Kim DW, Lee SH, Kim BJ. Naturally occurring mutations in large surface genes related to occult infection of hepatitis B virus genotype C. PLoS One. 2013; 8:e54486.

16). Kim HJ, Park JH, Jee Y, Lee SA, Kim H, Song BC, et al. Hepatitis B virus X mutations occurring naturally associated with clinical severity of liver disease among Korean patients with chronic genotype C infection. J Med Virol. 2008; 80:1337–43.

17). Lee SA, Cho YK, Lee KH, Hwang ES, Kook YH, Kim BJ. Gender disparity in distribution of the major hydrophilic region variants of hepatitis B virus genotype C according to hepatitis B e antigen serostatus. J Med Virol. 2011; 83:405–11.

18). Lee SA, Kim K, Kim H, Kim BJ. Nucleotide change of codon 182 in the surface gene of hepatitis B virus genotype C leading to truncated surface protein is associated with progression of liver diseases. J Hepatol. 2012; 56:63–9.

19). Lee SA, Kim KJ, Kim DW, Kim BJ. Male-specific W4P/R mutation in the pre-S1 region of hepatitis B virus, increasing the risk of progression of liver diseases in chronic patients. J Clin Microbiol. 2013; 51:3928–36.

20). Lee SA, Mun HS, Kim H, Lee HK, Kim BJ, Hwang ES, et al. Naturally occurring hepatitis B virus X deletions and insertions among Korean chronic patients. J Med Virol. 2011; 83:65–70.

21). Mun HS, Lee SA, Jee Y, Kim H, Park JH, Song BC, et al. The prevalence of hepatitis B virus preS deletions occurring naturally in Korean patients infected chronically with genotype C. J Med Virol. 2008; 80:1189–94.

22). Mun HS, Lee SA, Kim H, Hwang ES, Kook YH, Kim BJ. Novel F141L pre-S2 mutation in hepatitis B virus increases the risk of hepatocellular carcinoma in patients with chronic genotype C infections. J Virol. 2011; 85:123–32.

23). Song BC, Kim H, Kim SH, Cha CY, Kook YH, Kim BJ. Comparison of full length sequences of hepatitis B virus isolates in hepatocellular carcinoma patients and asymptomatic carriers of Korea. J Med Virol. 2005; 75:13–9.

24). Song BC, Kim SH, Kim H, Ying YH, Kim HJ, Kim YJ, et al. Prevalence of naturally occurring surface antigen variants of hepatitis B virus in Korean patients infected chronically. J Med Virol. 2005; 76:194–202.

25). Kim BJ. Hepatitis B virus mutations related to liver disease progression of Korean patients. World J Gastroenterol. 2014; 20:460–67.

26). Hosono S, Tai PC, Wang W, Ambrose M, Hwang DG, Yuan TT, et al. Core Antigen Mutations of Human Hepatitis B Virus in Hepatomas Accumulate in MHC Class II-Restricted T Cell Epitopes. Virology. 1995; 212:151–62.

27). Kim DW, Lee SA, Hwang ES, Kook YH, Kim BJ. Naturally occurring precore/core region mutations of hepatitis B virus genotype C related to hepatocellular carcinoma. PLoS One. 2012; 7:e47372.

28). Kim DW, Lee SA, Kim H, Won YS, Kim BJ. Naturally Occurring Mutations in the Nonstructural Region 5B of Hepatitis C Virus (HCV) from Treatment-Naïve Korean Patients Chronically Infected with HCV Genotype 1b. PLoS One. 2014; 9:e87773.

29). Wingfield PT, Stahl SJ, Williams RW, Steven AC. Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry. 1995; 34:4919–32.
30). Zhou S, Standring DN. Hepatitis B virus capsid particles are assembled from core-protein dimer precursors. Proc Natl Acad Sci U S A. 1992; 89:10046–50.

31). Bozkaya H, Ayola B, Lok AS. High rate of mutations in the hepatitis B core gene during the immune clearance phase of chronic hepatitis B virus infection. Hepatology. 1996; 24:32–7.

32). Kim D, Lyoo KS, Smith D, Hur W, Hong SW, Sung PS, et al. Number of mutations within CTL-defined epitopes of the hepatitis B Virus (HBV) core region is associated with HBV disease progression. J Med Virol. 2011; 83:2082–7.

34). Lim SG, Cheng Y, Guindon S, Seet BL, Lee LY, Hu P, et al. Viral quasi-species evolution during hepatitis Be antigen seroconversion. Gastroenterology. 2007; 133:951–8.

35). Ferrari E, He Z, Palermo RE, Huang HC. Hepatitis C Virus NS5B Polymerase Exhibits Distinct Nucleotide Requirements for Initiation and Elongation. J Biol Chem. 2008; 283:33893–901.

36). Hamano K, Sakamoto N, Enomoto N, Izumi N, Asahina Y, Kurosaki M, et al. Mutations in the NS5B region of the hepatitis C virus genome correlate with clinical outcomes of interferon-alpha plus ribavirin combination therapy. J Gastroenterol Hepatol. 2005; 20:1401–9.

37). Kim YJ, Lee JH, Kim YH. Studies on the Development of Viral Detection Markers for the Quality Control of Blood. J Bacteriol Virol. 2007; 37:177–91.

Go to : 
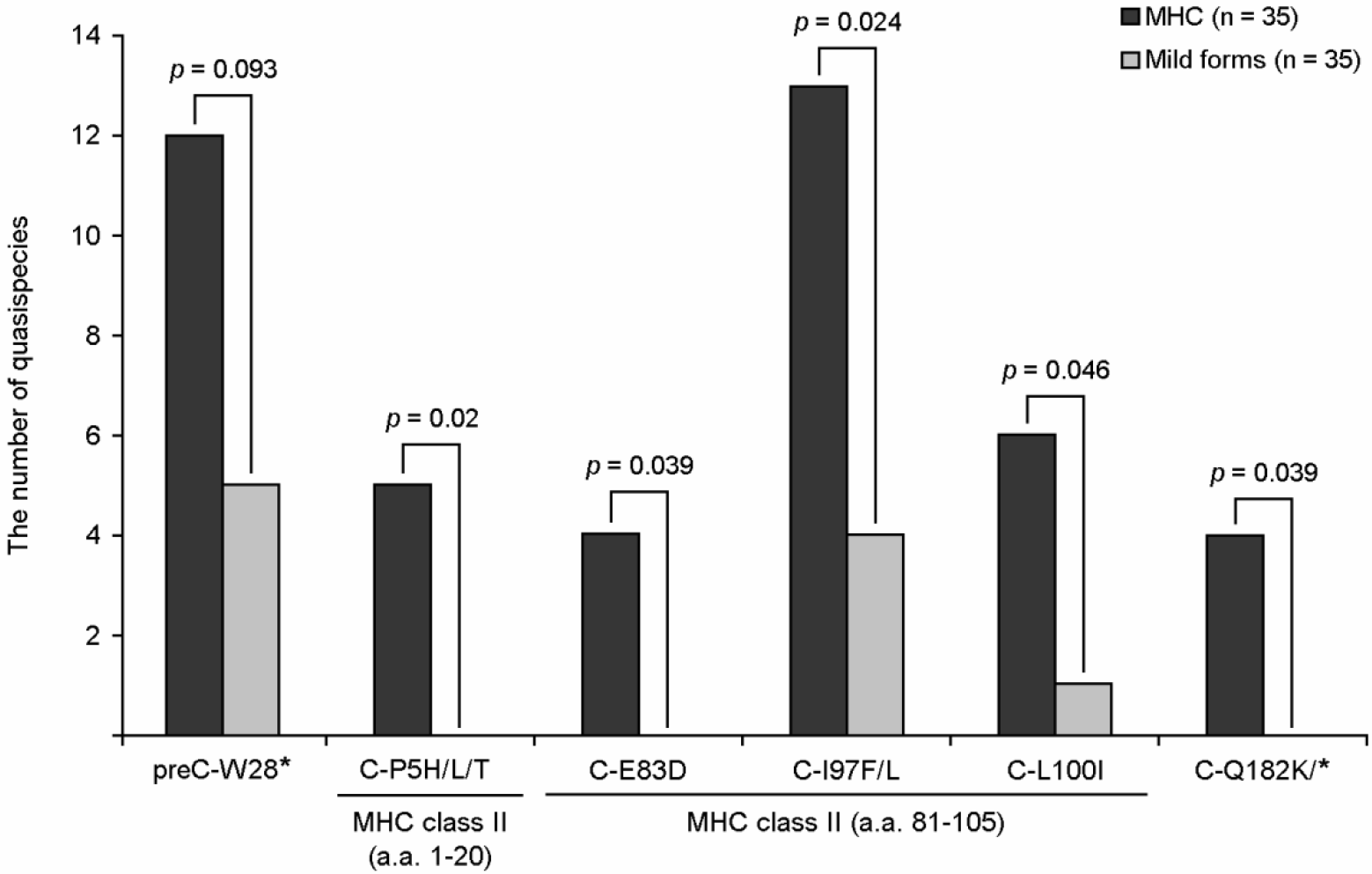 | Figure 1.
Six types of HBV preC/C mutations related to the severity of liver diseases in Korean chronic patients. The types of mutations within the region of MHC II-restricted T-cell epitopes are marked. For comparison between patients with HCC and other disease types, preC/C mutations of 35 HCC patients and 35 patients with other types of liver diseases (27 of chronic hepatitis and 8 patients of liver cirrhosis) were analyzed. To analyze the mutation patterns and their frequencies of deletions and insertions in the entire preC/C region, a nested PCR protocol was used. First-round PCR was performed using the sense primer CoreF1 (59-AAC GAC CGA CCT TGA GGC ATA CTT-39) and the antisense primer CoreR1 (59-ATT TGG TAA GGT TAG GAT AGA A-39) to yield a 1,017 bp amplicon between 1,682 nt to 2,698 nt of the HBV genome. Second-round PCR was performed using the sense primer CoreF2 (59-GAG TTG GGG GAG GAG ATT AGG TTA-39) and the antisense primer [11]. |
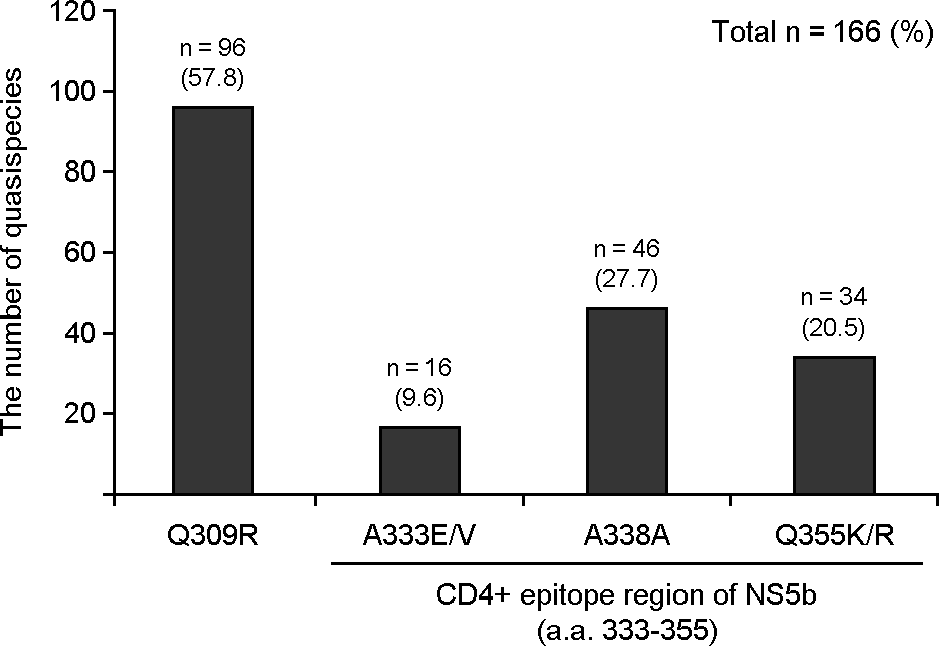 | Figure 2.
Mutation frequency at the 309, 333, 338 and 355 codons related to SVR and ETR in the NS5B region. The 166 quasispecies clones of HCV NS5B from 15 genotype 1b infected Korean patients were analyzed. For quasispecies analysis, the nested PCR protocol was performed. The first round of amplification was carried out using the sense primer A1b (O/S) (accession no. M62321, positions 8113-8135 59 – CTGACRACTAGCTGYGGTAAYAC – 39) and the antisense primer F1b (O/A) (positions 8678-8699, 59 – CCTGGAGAGTAACTRTGGAGTG – 39). The second round of amplification was carried out using the sense primer B1b (I/S) (positions 8181-8205, 59 – GCTCCRGGACTGCACSATGCTCGTG – 39) and the antisense primer E1b (I/A) (positions 8654-8675, 59 – AATGCGCTRAGRCCATGGAGTC – 39) to amplify 495 bp of the GT-1b NS5B region [28]. |
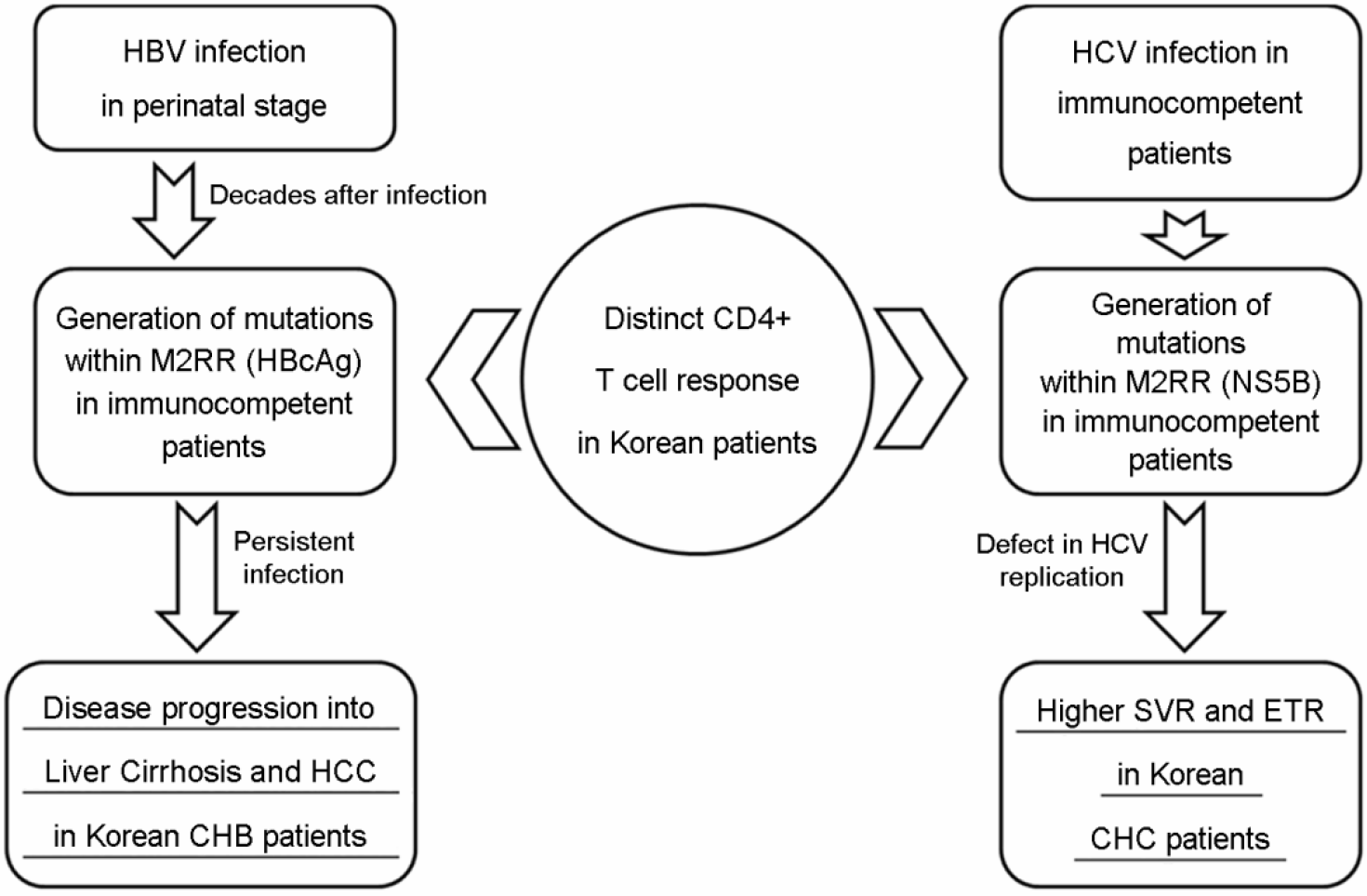 | Figure 3.
The presence of distinct CD4 T cell response in Korean patients may lead to opposite clinical outcomes in CHB and CHC Korean patients. The difference in clinical outcomes by viral mutations via distinct immune pressure between patients with CHB and CHC may be mainly due to the difference between HBV and HCV in the infections of Korean chronic patients, such as the vertical infection from mother to child versus horizontal infection. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download