Abstract
Helicobacter pylori, a gram-negative bacterium, is a causative agent of gastroduodenal diseases of human. Human immune system produces harmful reactive oxygen species to kill this bacterium that locates the microaerophilic mucous layer. H. pylori harbors various antioxidant enzymes including SodB, KatA and AhpC to protect the oxygen toxicity. We removed the catalase gene (katA) from H. pylori 26695 genome, and the change of profile of the gene expression of the mutant was analyzed by high resolution 2-DE followed by matrix assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS), tandem MS and microarray analysis. Eleven and 37 genes were upregulated and downregulated in the mutant respectively, either transcriptionally or translationally. Expression level of pfr and hp1588 that were decreased on protein level in the mutant was confirmed by RT-PCR analysis.
REFERENCES
1). Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984; 1:1311–5.

2). Nomura A, Stemmermann GN, Chyou PH, Perez-perez GI, Blaser MJ. Helicobacter pylori infection and the risk for duodenal and gastric ulceration. Ann Intern Med. 1994; 120:977–81.
3). Nomura A, Stemmermann GN, Chyou PH, Kato I, Perez-perez GI, Blaser MJ. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991; 325:1132–6.
4). No authors listed. Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7–14 June 1994. IARC monogr eval carcinog risks hum. 1994; 61:1–241.
5). Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999; 284:1328–33.
6). Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997; 388:539–47.
7). Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999; 397:176–80.
8). Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999; 21:33–7.

9). Seyler RW Jr, Olson JW, Maier RJ. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect Immun. 2001; 69:4034–40.
10). Ramarao N, Gray-Owen SD, Meyer TF. Helicobacter pylori induces but survives the extracellular release of oxygen radicals from professional phagocytes using its catalase activity. Mol Microbiol. 2000; 38:103–13.
11). Basu M1, Czinn SJ, Blanchard TG. Absence of catalase reduces long-term survival of Helicobacter pylori in macrophage phagosomes. Helicobacter. 2004; 9:211–6.
12). Wang G, Alamuri P, Maier RJ. The diverse antioxidant systems of Helicobacter pylori. Mol Microbiol. 2006; 61:847–60.
13). Cho MJ, Lee SG, Lee KH, Song JY, Lee WK, Baik SC, et al. Comparison of gene expression patterns between Helicobacter pylor 26695 and its superoxide dismutase isogenic mutant. J Bacteriol Virol. 2013; 43:279–89.
14). Alting-Mees MA, Short JM. pBluescript II: gene mapping vectors. Nucleic Acids Res. 1989; 17:9494.

15). Song JY, Park SG, Kang HL, Lee WK, Cho MJ, Park JU, et al. pHP489, a Helicobacter pylori small cryptic plasmid, harbors a novel gene coding for a replication initiation protein. Plasmid. 2003; 50:236–41.
16). Wang Y, Roos KP, Taylor DE. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J Gen Microbiol. 1993; 139:2485–93.
17). O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975; 250:4007–21.
18). Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976; 72:248–54.

19). Heukeshoven J, Dernick R. Improved silver staining procedure for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis. 1988; 9:28–32.

20). Park JW, Song JY, Hwang HR, Park HJ, Youn HS, Seo JH, et al. Proteomic analysis of thiol-active proteins of Helicobacter pylori 26695. J Bacteriol Virol. 2012; 42:211–23.
21). Chuang SE, Daniels DL, Blattner FR. Global regulation of gene expression in Escherichia coli. J Bacteriol. 1993; 175:2026–36.
22). Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997; 278:631–7.

23). Harris AG, Hinds FE, Beckhouse AG, Kolesnikow T, Hazell SL. Resistance to hydrogen peroxide in Helicobacter pylori: role of catalase (KatA) and Fur, and functional analysis of a novel gene product designated ‘KatA-associated protein’, KapA (HP0874). Microbiology. 2002; 148:3813–25.
24). Yamada H, Shimizu S, Shimada H, Tani Y, Takahashi S, Ohashi T. Production of D-phenylglycine-related amino acids by immobilized microbial cells. Biochimie. 1980; 62:395–9.

25). Baik SC, Kim KM, Song SM, Kim DS, Jun JS, Lee SG, et al. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of Helicobacter pylori strain 26695. J Bacteriol. 2004; 186:949–55.
26). Hochhauser SJ, Weiss B. Escherichia coli mutants deficient in deoxyuridine triphosphatase. J Bacteriol. 1978; 134:157–66.
27). Vicente JB, Ehrenkaufer GM, Saraiva LM, Teixeira M, Singh U. Entamoeba histolytica modulates a complex repertoire of novel genes in response to oxidative and nitrosative stresses: implications for amebic pathogenesis. Cell Microbiol. 2009; 11:51–69.
28). Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. Oxford: Clarendon Press;1989.
29). Slade D, Radman M. Oxidative Stress Resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev. 2011; 75:133–91.
30). Skouloubris S, Labigne A, De Reuse H. Identification and characterization of an aliphatic amidase in Helicobacter pylori. Mol Microbiol. 1997; 25:989–98.
31). Dalle-Donne I, Rossi R, Colombo R, Giustarini D, Milzani A. Biomarkers of oxidative damage in human disease. Clin Chem. 2006; 52:601–23.

32). Bereswill S, Waidner U, Odenbreit S, Lichte F, Fassbinder F, Bode G, et al. Structural, functional and mutational analysis of the pfr gene encoding a ferritin from Helicobacter pylori. Microbiology. 1998; 144:2505–16.
33). Nakamura A, Park A, Nagata K, Sato EF, Kashiba M, Tamura T, et al. Oxidative cellular damage associated with transformation of Helicobacter pylori from a bacillary to a coccoid form. Free Radic Biol Med. 2000; 28:1611–8.
34). Wang G, Maier RJ. An NADPH quinone reductase of Helicobacter pylori plays an important role in oxidative stress resistance and host colonization. Infect Immun. 2004; 72:1391–6.
35). Waidner B, Greiner S, Odenbreit S, Kavermann H, Velayudhan J, Stähler F, et al. Essential role of ferritin Pfr in Helicobacter pylori iron metabolism and gastric colonization. Infect Immun. 2002; 70:3923–9.
Figure 4.
Comparison of normalized gel images of wild type and kat isogenic mutant. The proteins were separated on an IPG strip of pH 5.0∼8.0 and subsequently on a 12.5% SDS-PAGE and then detected by silver staining. The original gel size was 18×20×0.15. (A) Wild type. (B) kat isogenic mutant.
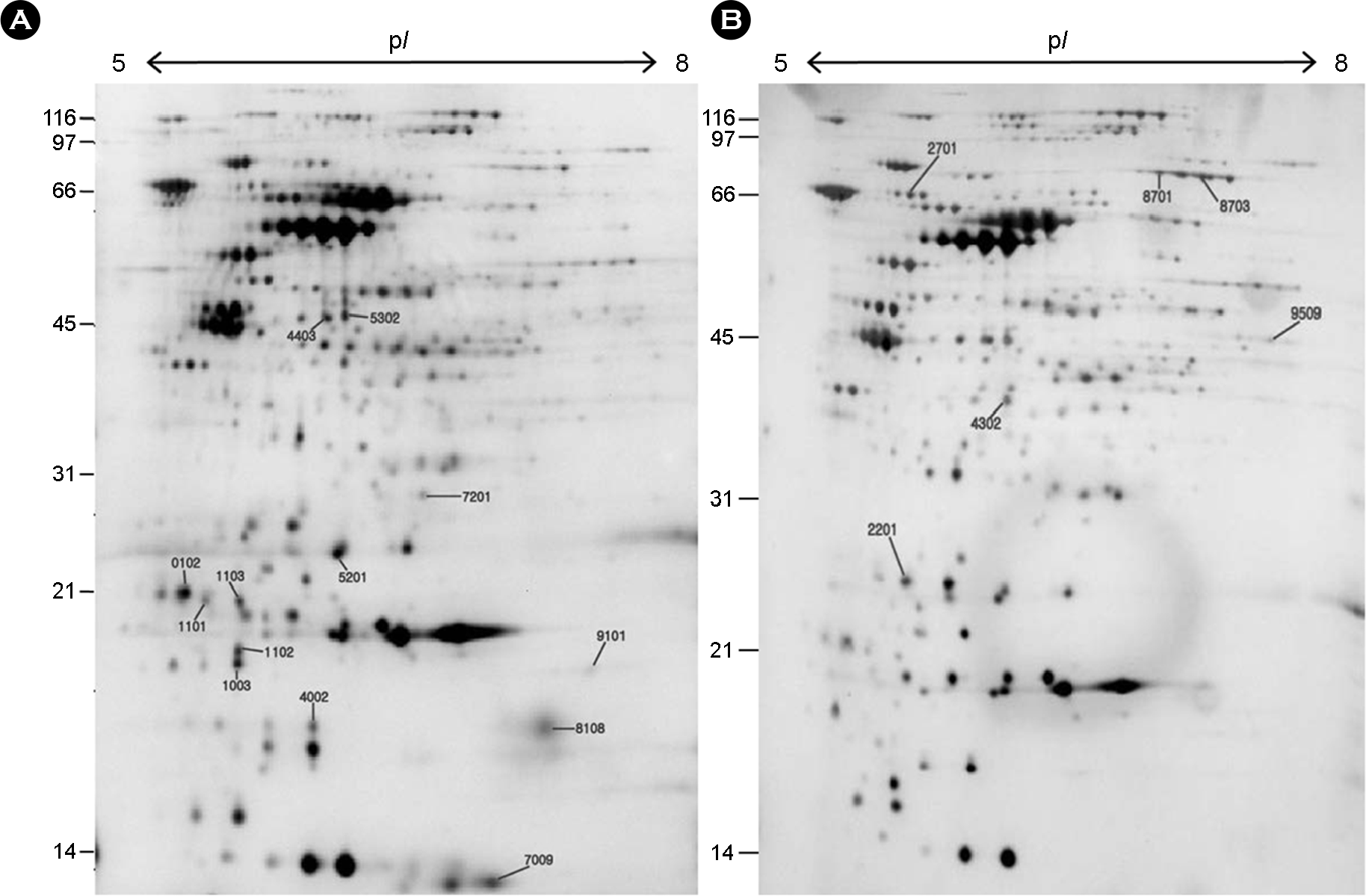
Figure 5.
Comparison of expression of genes in wild type H. pylori and in katA mutant. (A) RT-PCR analysis of pfr and hp1588. (B, C) expression of two genes. Levels were measured by 2-DE, microarray and RT-PCR.
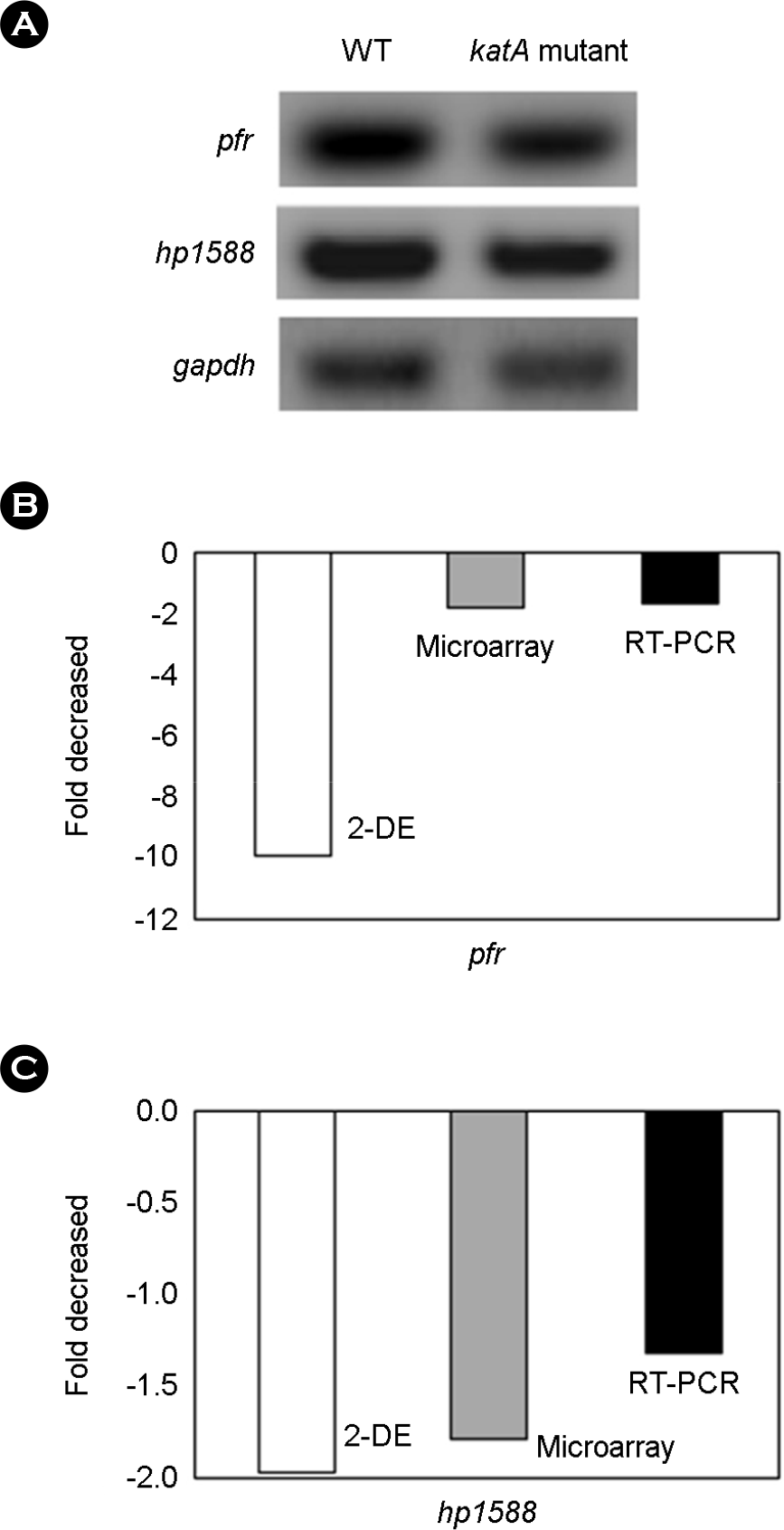
Table 1.
Strains and plasmids used in this study
Table 2.
List of up or down-regulated proteins of katA isogenic mutant of H. pylori 26695 by using tandem MS
| SSP no | Protein name | Accession no | mw | pI | Amino acid seq | Score | HP no |
|---|---|---|---|---|---|---|---|
| 0102b | Urease accessory protein (UreG) | gi:15644698 | 21,824 | 4.7 |
R.EAFNLIFKPGFSTAK.V R.AKEGLDDVIAWIK.R K.IDLAPYVGADLK.V |
45 33 55 |
0068 |
| 0903b | Histidine kinase (CheA) | gi:15645020 | 89,704 | 4.7 |
K.NGDKIPDAILVDIEMPK.M K.ITPDIMDVVLR.S K.VNITTLMNESENTK.S |
26 26 27 |
0903 |
| 1003b | Biotin carboxyl carrier protein (FabE) | gi:15644999 | 17,122 | 5.2 |
K.VVSVEVGDAQPVEYGTK.L K.EDFVLSPMVGTFYHAPSPGAEPYVK.V |
75 19 |
0371 |
| 1101b | Adenylate kinase (Adk) | gi:15645243 | 21,230 | 5.0 |
K.GIILIDGYPR.S R.VFLDPLGEIQNFYK.N |
39 53 |
0618 |
| 1102b | Biotin carboxyl carrier protein (FabE) | gi:15644999 | 17,122 | 5.2 |
K.VVSVEVGDAQPVEYGTK.L K.EDFVLSPMVGTFYHAPSPGAEPYVK.V |
64 44 |
0371 |
| 1103b | Adenylate kinase (Adk) | gi:15645243 | 21,230 | 5.0 |
K.ADMVEVFNFR.V K.GIILIDGYPR.S R.VFLDFLGEIQNFYK.N |
37 49 66 |
0618 |
| 2201a | Hydrogenase expression/formation protein (HypB) | gi:15645518 | 27,179 | 5.2 |
R.GTLFINPQTK.V K.TTMLENLADFK.D K.EGLYVLNFMSSPGSGK.T |
19 17 68 |
0900 |
| 2701a | GTP-binding protein, fusA-homolog (YihK) | gi:15645108 | 66,634 | 5.0 |
K.ALADEITLK.I K.INIIDTPGHADFGGEVER K.QLDFPVVYAAAR.D |
30 42 55 |
0480 |
| 4302a | Recombinase A (RecA) | gi:15644782 | 37,555 | 5.5 |
R.GLSLAGNQVLTR.T R.SGGIDLVVVDSVAALTPK.A K.AEIDGDMGDQHVGLQAR.L |
63 59 75 |
0153 |
| 5201b | Hypothetical protein HP1588 | gi:15646195 | 28,287 | 5.2 |
K.LLPGNEVIGPAIVESDATTFVIPK.G R.QALSAATLTLFK.M R.SSSQLLYSEIIVAGR.V |
33 46 67 |
1588 |
| 8701a | Hydantoin utilization protein A (HyuA) | gi:15645318 | 78,483 | 6.9 |
R.GVVATQKPVIPVEK.E R.TIVSGPIGGVIGSK.L R.LVLSLPLVAMDSVGAGAGSFVR.I K.IIQDAWDELTLK.V |
24 31 26 24 |
0695 |
| 8703a | Hydantoin utilization protein A (HyuA) | gi:15645318 | 78,483 | 6.9 |
K.YDDPLIPLKR.I R.TIVSGPIGGVIGSK.L |
44 59 |
0695 |
Table 3.
List of up or down-regulated proteins under katA isogenic mutant of H. pylori 26695 by using MALDI-TOF
| SSP no | Protein name | Accession no | mw | pI | HP no |
|---|---|---|---|---|---|
| 4002b | Nonheme iron-containing ferritin (Pfr) | gi:15645277 | 19,155 | 5.49 | 0653 |
| 4403b | Pyruvate: ferredoxin oxidoreductase, alpha Subunit (PorA) | gi:15645724 | 44,613 | 5.81 | 1110 |
| 5302b | GTP-binding protein (Gtp1) | gi:15645194 | 40,444 | 5.53 | 0569 |
| 7009b | Co-chaperone (GroES) | gi:15644644 | 12,860 | 6.59 | 0011 |
| 8108b | Adhesin-thiol peroxidase (TagD) | gi:15645018 | 18,161 | 8.18 | 0390 |
| 9509a | Fumarase (FumC) | gi:15645938 | 50,844 | 7.29 | 1325 |
Table 4.
List of genes whose expression was increased over two-fold in kat isogenic mutant compared with wild type H. pylori 26695
| HP No. | Increasing level (fold)* | Gene name |
|---|---|---|
| HP0686 | 2.018 | Iron (III) dicitrate transport protein (fecA) |
| HP0810 | 2.056 | Conserved hypothetical protein |
| HP0865 | 2.452 | Deoxyuridine 5′-triphosphate nucleotidohydrolase (dut) |
| HP0870 | 2.457 | Flagellar hook (flgE) |
| HP0871 | 3.109 | CDP-diglyceride hydrolase (cdh) |
| HP1174 | 2.982 | Glucose/galactose transporter (gluP) |
Table 5.
List of genes whose expression was decreased over twofold in katA isogenic mutant compared with wild type H. pylori 26695
| HP No. | Decreasing level (fold)* | Gene name |
|---|---|---|
| HP0004 | 0.392 | Carbonic anhydrase (icfA) |
| HP0102 | 0.330 | Conserved hypothetical protein |
| HP0119 | 0.431 | Hypothetical protein |
| HP0373 | 0.481 | Conserved hypothetical protein |
| HP0408 | 0.450 | Hypothetical protein |
| HP0415 | 0.463 | Conserved hypothetical integral membrane protein |
| HP0439 | 0.442 | Hypothetical protein |
| HP0503 | 0.162 | Hypothetical protein |
| HP0513 | 0.431 | Hypothetical protein |
| HP0653 | 0.543 | Nonheme iron-containing ferritin (Pfr) |
| HP0587 | 0.377 | Aminodeoxychorismate lyase (pabC) |
| HP0600 | 0.351 | Multidrug resistance protein (spaB) |
| HP0603 | 0.469 | Hypothetical protein |
| HP0614 | 0.466 | Hypothetical protein |
| HP0689 | 0.397 | Hypothetical protein |
| HP0702 | 0.365 | Hypothetical protein |
| HP0704 | 0.440 | Hypothetical protein |
| HP0712 | 0.185 | Hypothetical protein |
| HP0767 | 0.499 | Hypothetical protein |
| HP0784 | 0.490 | Hypothetical protein |
| HP0889 | 0.397 | Iron (III) dicitrate ABC transporter permease protein (fecD) |
| HP0981 | 0.481 | Exonuclease VII-like protein (xseA) |
| HP1142 | 0.420 | Hypothetical protein |
| HP1227 | 0.477 | Cytochrome c553 |
| HP1238a | 0.421 | Aliphatic amidase (amiE) |
| HP1390 | 0.287 | Hypothetical protein |
| HP1426 | 0.478 | Conserved hypothetical protein |
| HP1534 | 0.469 | IS605 transposase (tnpB) |
| HP1588 | 0.577 | Hypothetical protein |
Table 6.
Clusters of orthologous groups of genes (COGs) of 32 genes 2-fold changes in expression levels of katA mutants compared with wild type H. pylori 26695.
Table 7.
Tentative annotation of 7 hypothetical proteins by the clustering analysis.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download