Abstract
Clostridium perfringens food poisoning ranks among the most common gastrointestinal diseases in developed countries. In Korea, C. perfringens food poisoning gradually increases. Using PCR, 72 strains of C. perfringens isolated in Seoul, 2013 were tested for the presence of toxin genes. Of the tested strains, 32 isolates carried the cpe gene, 37 isolates carried the cpb2 gene and 3 isolates carried the cpe and cpb2 genes, respectively. 32 cpe- positive strains were isolated from the food poisoning patient, whereas among 37 cpb2-positive strains, 22 strains were isolated from asymptomatic person. To investigate epidemiological relationship between the isolates, Pulsed-filed gel electrophoresis (PFGE) was performed. The genetic relatedness of the isolates ranged from 55.9% to 100% and 47 distinct PFGE profiles were observed. The results show that the cpe- positive outbreak strains showed close genetic relation, whereas the cpb2-positive isolates revealed a wide genetic diversity.
Go to : 
REFERENCES
1). Smedley JG 3rd, Fisher DJ, Sayeed S, Chakrabarti G, McClane BA. The enteric toxins of Clostridium perfringens. Rev Physiol Biochem Pharmacol. 2004; 152:183–204.
2). Sartory DP, Field M, Curbishly SM, Pritchard AM. Evaluation of two media for the membrane filtration enumeration of Clostridium perfringens from water. Lett Appl Microbiol. 1998; 27:323–7.
3). Sarker MR, Singh U, McClane BA. An update on Clostridium perfringens enterotoxin. J Nat Toxins. 2000; 9:251–66.
4). McDonel JL. Clostridium perfringens toxins (type A, B, C, D, E). Pharmacol Ther. 1980; 10:617–55.
5). Petit L, Gibert M, Popoff MR. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 1999; 7:104–10.
6). Harrison B, Raju D, Garmory HS, Brett MM, Titball RW, Sarker MR. Molecular characterization of Clostridium perfringens isolates from humans with sporadic diarrhea: evidence for transcriptional regulation of the beta2-toxin-encoding gene. Appl Environ Microbiol. 2005; 71:8362–70.
7). Fisher DJ, Miyamoto K, Harrison B, Akimoto S, Sarker MR, McClane BA. Association of beta2 toxin production with Clostridium perfringens type A human gastrointestinal disease isolates carrying a plasmid enterotoxin gene. Mol Microbiol. 2005; 56:747–62.
8). Sparks SG, Carman RJ, Sarker MR, McClane BA. Genotyping of enterotoxigenic Clostridium perfringens fecal isolates associated with antibiotic-associated diarrhea and food poisoning in North America. J Clin Microbiol. 2001; 39:883–8.
9). Songer JG. Clostridial enteric disease of domestic animals. Clin Microbiol Rev. 1996; 9:216–34.
10). Cornillot E, Saint-Joanis B, Daube G, Katayama S, Granum PE, Canard B, et al. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol Microbiol. 1995; 15:639–47.
11). McCabe-Sellers BJ, Beattie SE. Food safety: Emerging trends in foodborne illness surveillance and prevention. J Am Diet Assoc. 2004; 104:1708–17.

13). Chalmers G, Martin SW, Hunter DB, Prescott JF, Weber LJ, Boerlin P. Genetic diversity of Clostridium perfringens isolated from healthy broiler chickens at a commercial farm. Vet Microbiol. 2008; 127:116–27.
14). Chon JW, Park JS, Hyeon JY, Park C, Song KY, Hong KW, et al. Development of Real-time PCR for the detection of Clostridium perfringens in meats and vegetables. J Microbiol Biotechnol. 2012; 22:530–4.
15). Kim NO, Cha I, Kim JS, Chung GT, Kang YH, Hong S. The prevalence and characteristics of bacteria causing acute diarrhea in Korea, 2012. Ann Clin Microbiol. 2013; 16:174–81.

16). Rood JI, Cole ST. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol Rev. 1991; 55:621–48.
17). Collie RE, Kokai-Kun JF, McClane BA. Phenotypic characterization of enterotoxigenic Clostridium perfringens isolates from non-foodborne human gastrointestinal diseases. Anaerobe. 1998; 4:69–79.
18). Fisher DJ, Miyamoto K, Harrison B, Akimoto S, Sarker MR, McClane BA. Association of beta2 toxin production with Clostridium perfringens type A human gastrointestinal disease isolates carrying a plasmid enterotoxin genes. Mol Microbiol. 2005; 56:747–62.
Go to : 
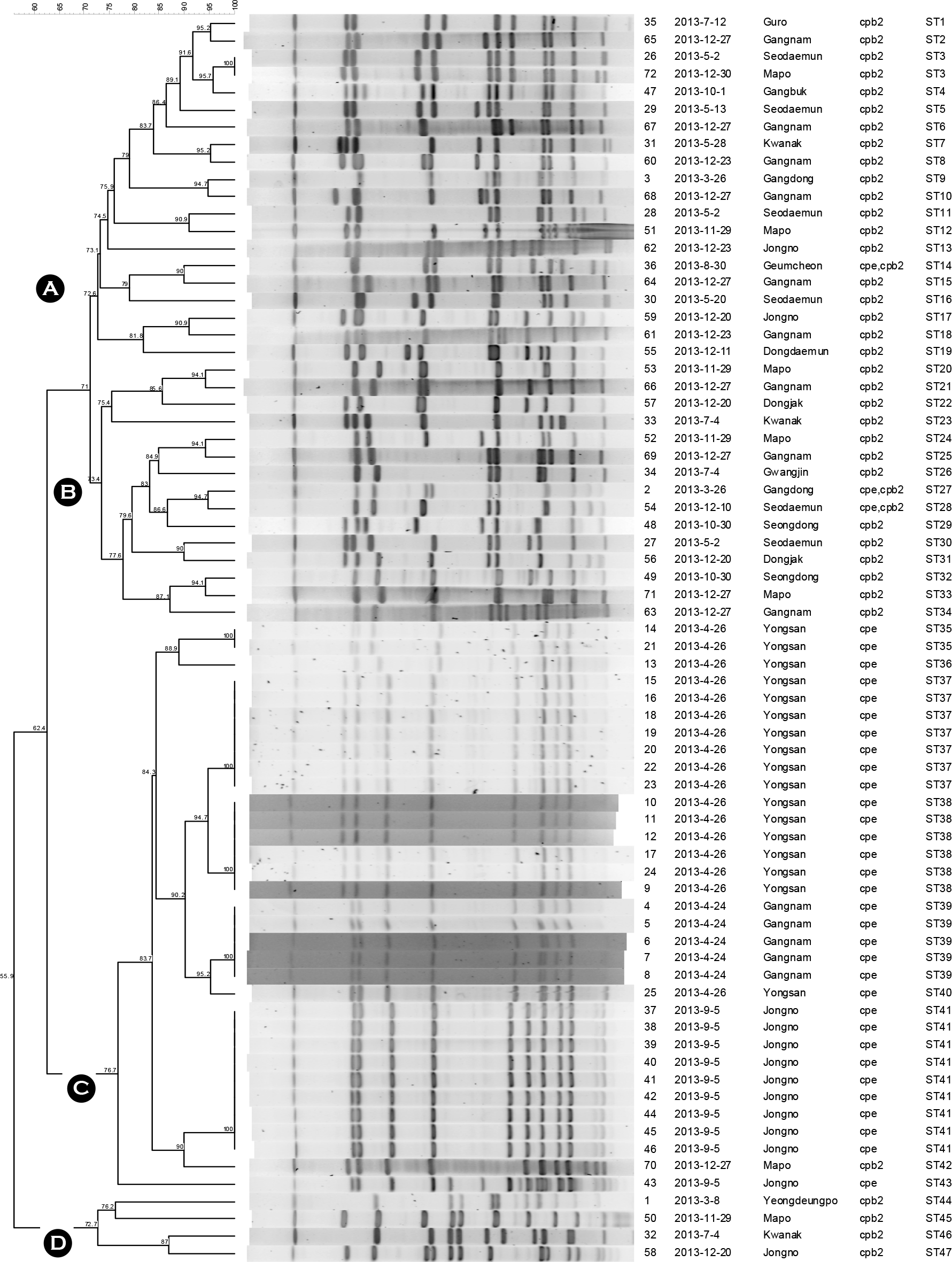 | Figure 1.Dendrogram showing the clustering of PFGE patterns for the 72 C. perfringens isolates. A group indicates 72.6% similarity; B group indicates 73.4% similarity; C group indicates 76.7% similarity; D group indicates 72.7% similarity. ST means PFGE subtype pattern. |
Table 1.
Isolation and region distribution of 72 C. perfringens isolated in Seoul, 2013




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download