Abstract
Oral infection with Porphyromonas (P.) gingivalis causes periodontitis that is manifested by the destruction of gingival connective tissues. Although a few types of antibiotics are effective against the infection, its use induces the appearance of drug-resistant bacteria. The present study shows that the fermented product of Aspergillus (A.) oryzae S-03, cultivated on the fat-removed soybean, inhibits the cell growth of the P. gingivalis. Likewise, the fermented product of the S-03 strain cultured for 26∼42 h displays an inhibitory activity to gingipain as a virulence factor of P. gingivalis. The activity is not lost even with heat treatment at 100°C for 15 min. We also demonstrate that the S-03 strain exhibits high protease activity. In addition, the strain does not produce aflatoxin because of the loss of a regulatory gene, aflR, necessary for the toxin biosynthesis.
Go to : 
REFERENCES
1). Abrar M, Anjum FM, Butt MS, Pasha I, Randhawa MA, Saeed F, et al. Aflatoxins: Biosynthesis, Occurrence, Toxicity and Remedies. Crit Rev Food Sci Nutr. 2013; 53:862–74.

2). Hesseltine CW, Shotwell OL, Ellis JJ, Stubblefield RD. Aflatoxin formation by Aspergillus flavus. Bacteriol Rev. 1966; 30:795–805.
3). Kew MC. Aflatoxins as a cause of hepatocellular carcinoma. J Gastrointestin Liver Dis. 2013; 22:305–10.
4). Bhanja T, Kumari A, Banerjee R. Enrichment of phenolics and free radical scavenging property of wheat koji prepared with two filamentous fungi. Bioresour Technol. 2009; 100:2861–6.

5). Hajjaj H, Duboc P, Fay LB, Zbinden I, Macé K, Niederberger P. Aspergillus oryzae produces compounds inhibiting cholesterol biosynthesis downstream of dihydrolanosterol. FEMS Microbiol Lett. 2005; 242:155–9.
6). Inoue K, Gotou T, Kitajima H, Mizuno S, Nakazawa T, Yamamoto N. Release of antihypertensive peptides in miso paste during its fermentation, by the addition of casein. J Biosci Bioeng. 2009; 108:111–5.

7). Gibson FC 3rd, Genco CA. Porphyromonas gingivalis mediated periodontal disease and atherosclerosis: disparate diseases with commonalities in pathogenesis through TLRs. Curr Pharm Des. 2007; 13:3665–75.
8). Griffen AL, Becker MR, Lyons SR, Moeschberger ML, Leys EJ. Prevalence of Porphyromonas gingivalis and periodontal health status. J Clin Microbiol. 1998; 36:3239–42.
9). Chen T, Nakayama K, Belliveau L, Duncan MJ. Porphyromonas gingivalis gingipains and adhesion to epithelial cells. Infect Immun. 2001; 69:3048–56.
10). Curtis MA, Aduse-Opoku J, Rangarajan M. Cysteine proteases of Porphyromonas gingivalis. Crit Rev Oral Biol Med. 2001; 12:192–216.
11). Imamura T. The role of gingipains in the pathogenesis of periodontal disease. J Periodontol. 2003; 74:111–8.

12). Potempa J, Mikolajczyk-Pawlinska J, Brassell D, Nelson D, Thøgersen IB, Enghild JJ, et al. Comparative properties of two cysteine proteinases (Gingipains R), the products of two related but individual genes of Porphyromonas gingivalis. J Biol Chem. 1998; 273:21648–57.
13). Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr Protein Pept Sci. 2003; 4:397–407.
14). Japoni A, Vasin A, Noushadi S, Kiany F, Japoni S, Alborzi A. Antibacterial susceptibility patterns of Porphyromonas gingivalis isolated from chronic periodontitis patients. Med Oral Patol Oral Cir Bucal. 2011; 16:1031–5.
15). Kadowaki T, Baba A, Abe N, Takii R, Hashimoto M, Tsukuba T, et al. Suppression of pathogenicity of Porphyromonas gingivalis by newly developed gingipain inhibitors. Mol Pharmacol. 2004; 66:1599–606.
16). Bedi GS. Comparative study of four proteases from spent culture media of Porphyromonas gingivalis (FAY-19M-1). Prep Biochem. 1995; 25:133–54.
17). Kitano S, Irimura K, Sasaki T, Abe N, Baba A, Miyake Y, Katunuma N, et al. Suppression of gingival inflammation induced Porphyromonas gingivalis in rats by leupeptin. Jpn J Pharmacol. 2001; 85:84–91.
18). Monti B, Sparapani M, Contestabile A. Differential toxicity of protease inhibitors in cultures of cerebellar granule neurons. Exp Neurol. 1998; 153:335–41.

19). Labrecque J, Bodet C, Chandad F, Grenier D. Effects of a high-molecular-weight cranberry fraction on growth, biofilm formation and adherence of Porphyromonas gingivalis. J Antimicrob Chemother. 2006; 58:439–43.
20). Okamoto M, Sugimoto A, Leung KP, Nakayama K, Kamaguchi A, Maeda N. Inhibitory effect of green tea catechins on cysteine proteinases in Porphyromonas gingivalis. Oral Microbiol Immunol. 2004; 19:118–20.
21). Sakanaka S, Aizawa M, Kim M, Yamamoto T. Inhibitory effects of green tea polyphenols on growth and cellular adherence of an oral bacterium, Porphyromonas gingivalis. Biosci Biotechnol Biochem. 1996; 60:745–9.
22). Toh EC, Dashper SG, Huq NL, Attard TJ, O'Brien-Simpson NM, Chen YY, et al. Porphyromonas gingivalis cysteine proteinase inhibition by κ-casein peptides. Antimicrob Agents Chemother. 2011; 55:1155–61.
23). Miyasaki KT, Iofel R, Oren A, Huynh T, Lehrer RI. Killing of Fusobacterium nucleatum, Porphyromonas gingivalis and Prevotella intermedia by protegrins. J Periodontal Res. 1998; 33:91–8.
24). Wang HL, Ellis JJ, Hesseltine CW. Antibacterial activity produced by molds commonly used in oriental food fermentations. Mycologia. 1972; 64:218–21.

25). Jin J, Lee YK, Wickes BL. Simple chemical extraction method for DNA isolation from Aspergillus fumigatus and other Aspergillus species. J Clin Microbiol. 2004; 42:4293–6.
26). Potempa J, Nguyen KA. Purification and characterization of gingipains. Curr Protoc Protein Sci. 2007.

27). Henry T, Iwen PC, Hinrichs SH. Identification of Aspergillus species using internal transcribed spacer regions 1 and 2. J Clin Microbiol. 2000; 38:1510–5.
28). Criseo G, Bagnara A, Bisignano G. Differentiation of aflatoxin-producing and non-producing strains of Aspergillus flavus group. Lett Appl Microbiol. 2001; 33:291–5.
29). El-Hag N, Morse RE. Aflatoxin production by a variant of Aspergillus oryzae (NRRL strain 1988) on cowpeas (Vigna sinensis). Science. 1976; 192:1345–6.
30). Tominaga M, Lee YH, Hayashi R, Suzuki Y, Yamada O, Sakamoto K, et al. Molecular analysis of an inactive aflatoxin biosynthesis gene cluster in Aspergillus oryzae RIB strains. Appl Environ Microbiol. 2006; 72:484–90.
31). Ehrlich KC, Yu J, Cotty PJ. Aflatoxin biosynthesis gene clusters and flanking regions. J Appl Microbiol. 2005; 99:518–27.

32). Kiyota T, Hamada R, Sakamoto K, Iwashita K, Yamada O, Mikami S. Aflatoxin non-productivity of Aspergillus oryzae caused by loss of function in the aflJ gene product. J Biosci Bioeng. 2011; 111:512–7.
33). Machida M, Yamada O, Gomi K. Genomics of Aspergillus oryzae: learning from the history of Koji mold and exploration of its future. DNA Res. 2008; 15:173–83.
34). Payne GA, Nystrom GJ, Bhatnagar D, Cleveland TE, Woloshuk CP. Cloning of the afl-2 gene involved in aflatoxin biosynthesis from Aspergillus flavus. Appl Environ Microbiol. 1993; 59:156–62.
35). Yu JH, Butchko RA, Fernandes M, Keller NP, Leonard TJ, Adams TH. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr Genet. 1996; 29:549–55.
36). Kim EY, Rhyu MR. Antimicrobial activities of Monascus koji extracts. Korean J Food Sci Technol. 2008; 40:76–81.
37). Ungureanu C, Ferdes M. Antibacterial and antifungal activity of red rice obtained from Monascus purpureus. Chem Eng. 2010; 20:223–8.
Go to : 
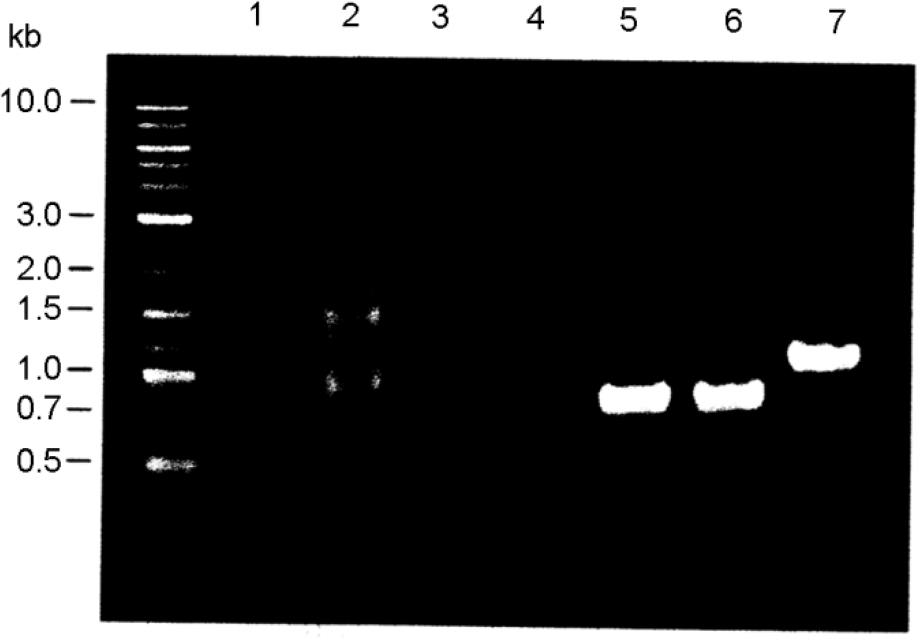 | Figure 1.
PCR amplification specificity of the genomic DNA of A. oryzae S-03 using primers designed for the aflatoxin gene cluster. The primer sequences are shown in Table 1. Lanes: 1, aflT; 2, nor-1; 3, aflR; 4, norA; 5, avnA; 6, verB; 7, vbs
|
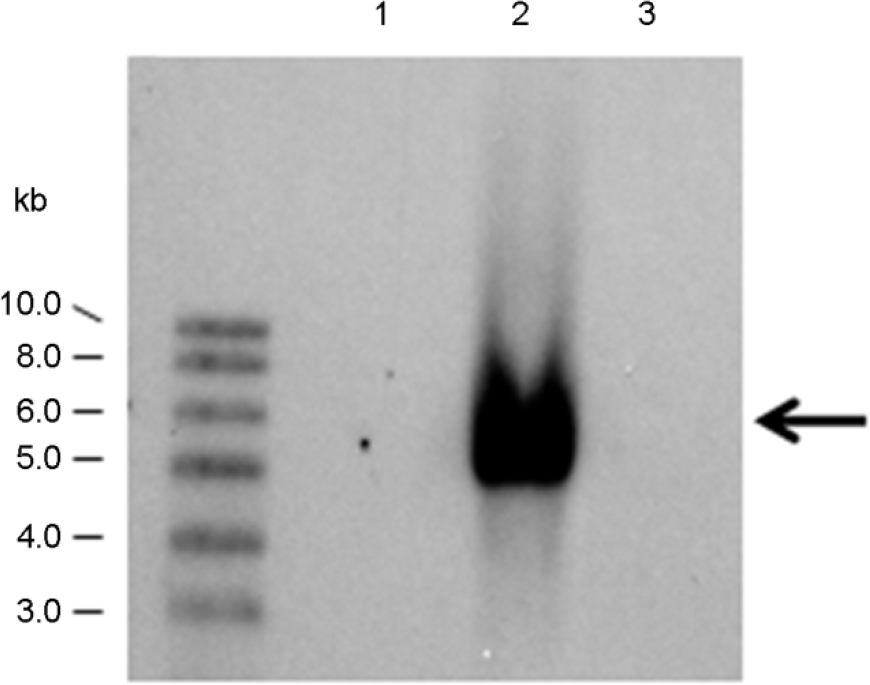 | Figure 2.
Southern blot analysis of the PstI-digested chromosomal DNA from Aspergillus species. The aflR fragment was used as a probe. The arrow indicates the molecular mass of the signals. Lanes 1, A. oryzae M-01; 2, A. oryzae W-52; 3, A. oryzae S-03. The M-01 and W-52 strains are classified into group 2 and group 1, respectively. The aflR gene found in the W-52 strain, but not in the M-01 and S-03 strains. |
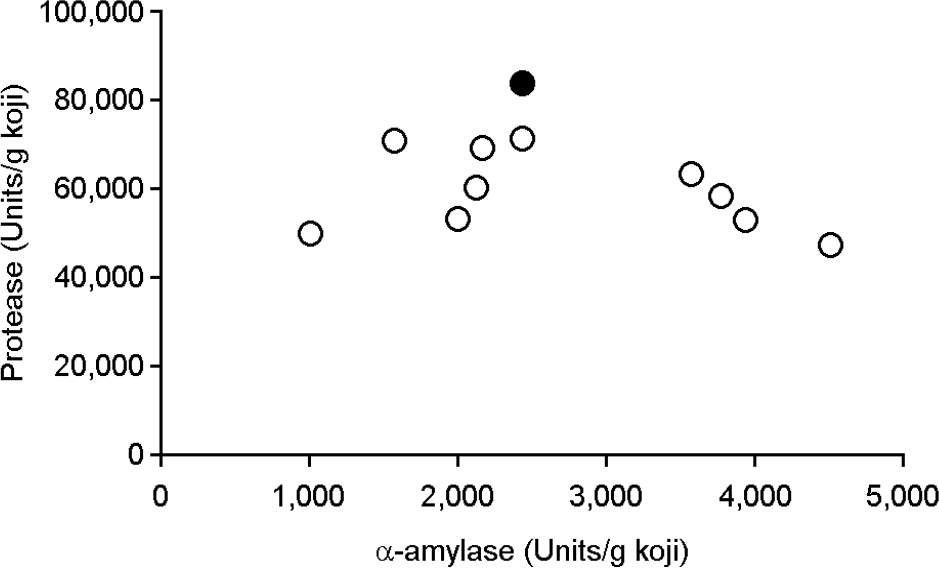 | Figure 3.
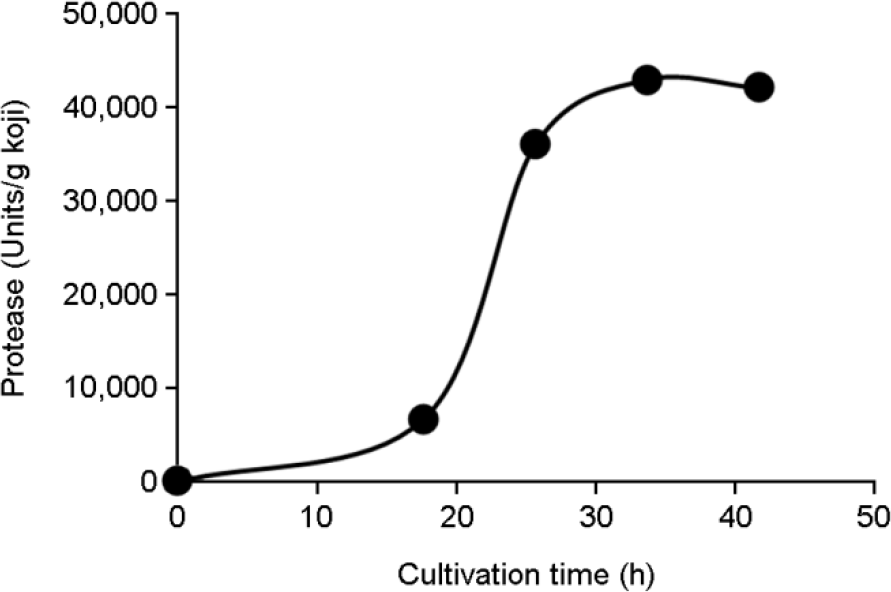
Activities of α-amylase and protease produced by A. oryzae. The detailed measurement method is described in the Materials and Methods section. Hatched rectangle, A. oryzae S-03. |
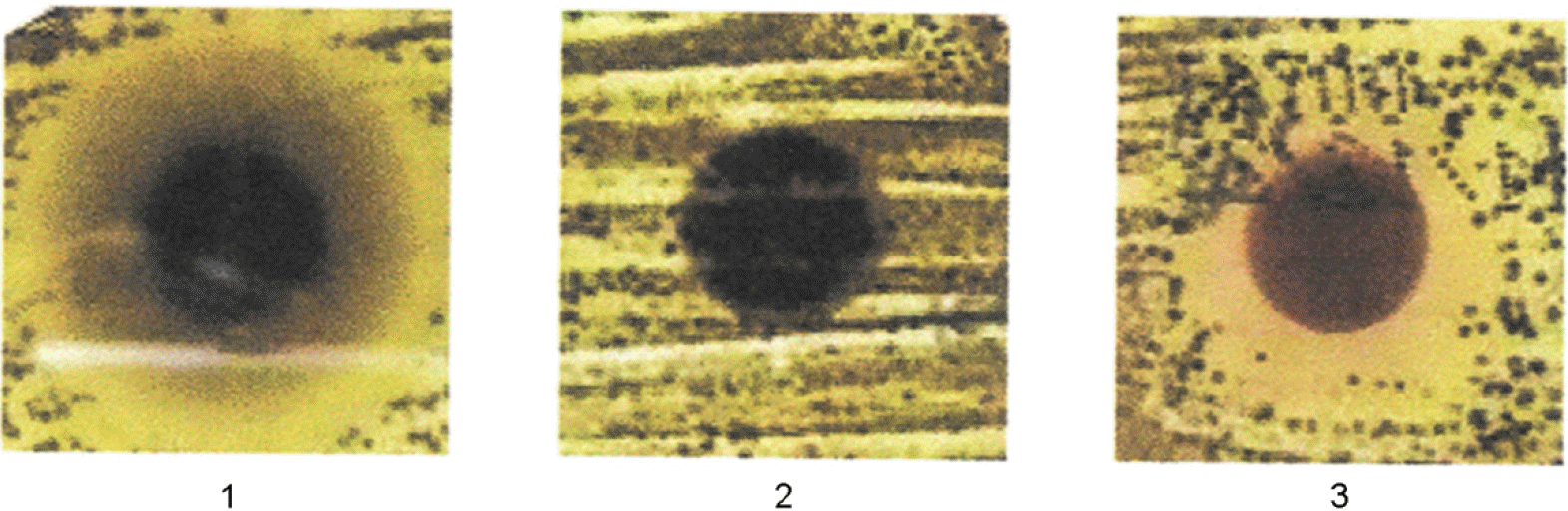 | Figure 5.
Antibacterial activity produced by A. oryzae S-03 cultivated on the fat-removed soybeans. The supernatant fluid was obtained extraction of koji and applied on paper disc. 1, non-heat treated supernatant fluid; 2, the fluid treated with 100°C for 10 min; 3, the fluid treated with 60°C for 30 min. |
Table 1.
Primers used in this study
Table 2.
Activity of Rgp and Kgp proteases of P. gingivalis W83. The values are expressed as percentage of the activity determined in the absence of inhibitor.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download