Abstract
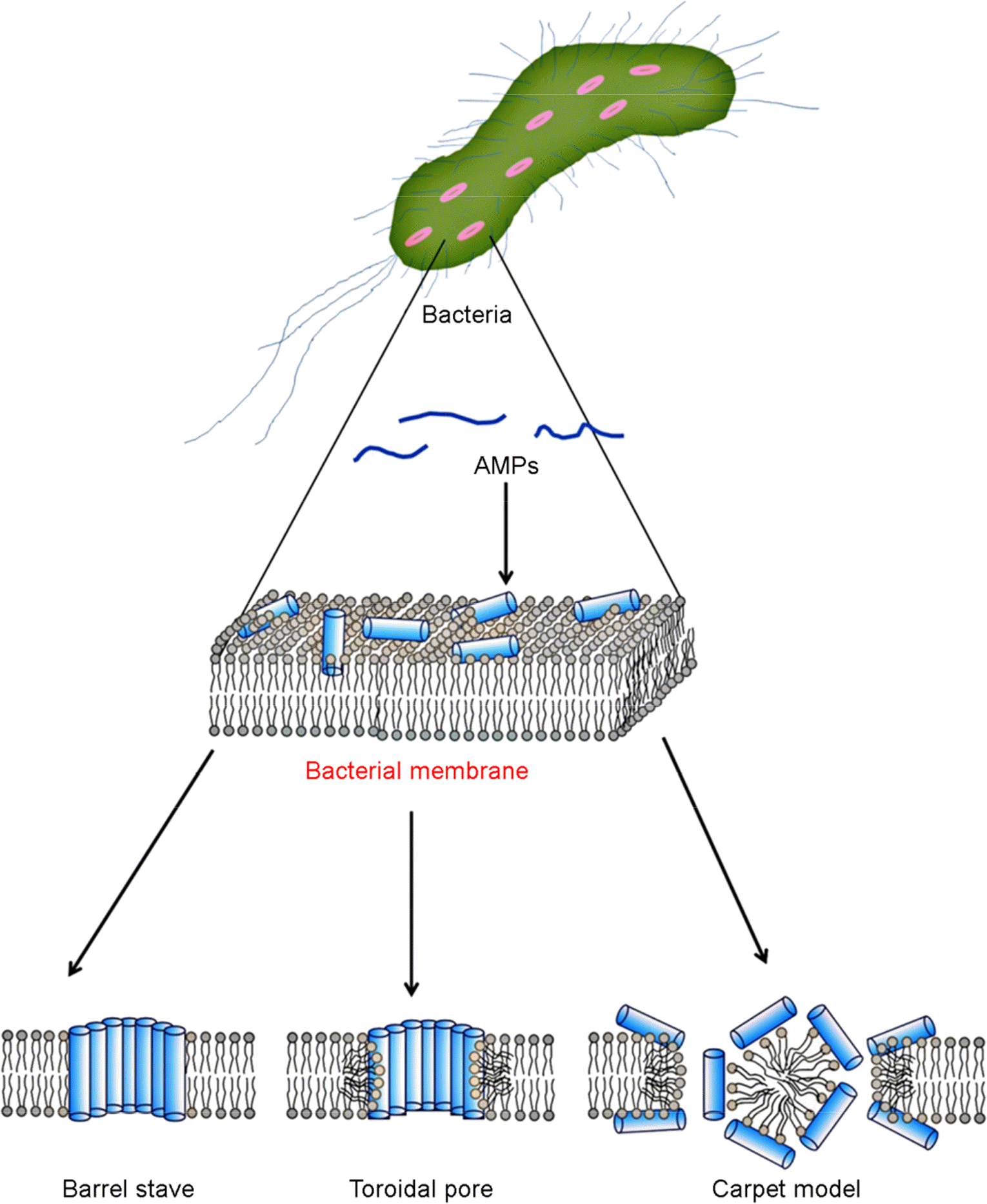
Resistance to antibiotics is becoming a very serious problem, with so-called superbugs exhibiting resistance to nearly all conventional antibiotic drugs. Consequently, these organisms often cause severe illness and even death. Alternatives to conventional antibiotics are antimicrobial peptides (AMPs). These widely expressed short peptides, which have been isolated from insects, plants, marine organisms and mammals, including humans, show strong antimicrobial activity against both Gram-negative and Gram-positive bacteria. Most AMPs act by disrupting the bacterial membrane through “Barrel-stave”, “Toroidal pore”, “carpet” mechanism. In addition, AMPs may prevent septic shock through strongly binding lipopolysaccharides and lipoteichoic acid located on the bacterial membrane. The action mechanisms of AMP to minimize the likelihood developing resistance to the peptides would be particular advantage. For these reasons, we anticipate that AMPs will replace conventional antibiotic drugs in a variety of contexts.
Go to : 
REFERENCES
1). Zhang QG. Exposure to phages has little impact on the evolution of bacterial antibiotic resistance on drug concentration gradients. Evol Appl. 2014; 7:394–402.

2). Grummet JP, Weerakoon M, Huang S, Lawrentschuk N, Frydenberg M, Moon DA, et al. Sepsis and ‘superbugs’: should we favour the transperineal over the transrectal approach for prostate biopsy? BJU Int. 2013.

3). Nordmann P, Naas T, Fortineau N, Poirel L. Superbugs in the coming new decade; multidrug resistance and prospects for treatment of Staphylococcus aureus, Enterococcus spp. and Pseudomonas aeruginosa in 2010. Curr Opin Microbiol. 2007; 10:436–40.
4). Xu ZQ, Flavin MT, Flavin J. Combating multidrug-resistant Gram-negative bacterial infections. Expert Opin Investig Drugs. 2014; 23:163–82.

5). Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 2009; 5:e1000354.
6). Adav SS, Lin JC, Yang Z, Whiteley CG, Lee DJ, Peng XF, et al. Stereological assessment of extracellular polymeric substances, exo-enzymes, and specific bacterial strains in bioaggregates using fluorescence experiments. Biotechnol Adv. 2010; 28:255–80.

7). Tsai MW, Lee DJ, Lai JY. Mass transfer limit of fluorescent dyes during multicolor staining of aerobic granules. Appl Microbiol Biotechnol. 2008; 78:907–13.

8). Roscia G, Falciani C, Bracci L, Pini A. The development of antimicrobial peptides as new antibacterial drugs. Curr Protein Pept Sci. 2013; 14:641–9.

9). Guilhelmelli F, Vilela N, Albuquerque P, Derengowski LD, Silva-Pereira I, Kyaw CM. Antibiotic development challenges: the various mechanisms of action of antimicrobial peptides and of bacterial resistance. Front Microbiol. 2013; 4:353.

11). Lee JK, Ramamourthy Gopal, Park Y. The function and application of antimicrobial peptides. Appl Chem Eng. 2011. 119–24.
12). Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005; 3:238–50.

13). Payne JW, Jakes R, Hartley BS. The primary structure of alamethicin. Biochem J. 1970; 117:757–66.

14). Hall JE, Vodyanoy I, Balasubramanian TM, Marshall GR. Alamethicin. A rich model for channel behavior. Biophys J. 1984; 45:233–47.

15). Rinehart KL Jr, Cook JC Jr, Meng H, Olson KL, Pandey RC. Mass spectrometric determination of molecular formulas for membrane-modifying antibiotics. Nature. 1977; 269:832–3.

16). Amiche M, Seon AA, Wroblewski H, Nicolas P. Isolation of dermatoxin from frog skin, an antibacterial peptide encoded by a novel member of the dermaseptin genes family. Eur J Biochem. 2000; 267:4583–92.

17). Wang KF, Nagarajan R, Camesano TA. Antimicrobial peptide alamethicin insertion into lipid bilayer: A QCM-D exploration. Colloids Surf B Biointerfaces. 2014; 116:472–81.

18). Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci U S A. 1987; 84:5449–53.

19). Matsuzaki K, Sugishita K, Harada M, Fujii N, Miyajima K. Interactions of an antimicrobial peptide, magainin 2, with outer and inner membranes of Gram-negative bacteria. Biochim Biophys Acta. 1997; 1327:119–30.

20). Matsuzaki K, Sugishita K, Fujii N, Miyajima K. Molecular basis for membrane selectivity of an antimicrobial peptide, magainin 2. Biochemistry. 1995; 34:3423–9.

21). Matsuzaki K, Nakayama M, Fukui M, Otaka A, Funakoshi S, Fujii N, et al. Role of disulfide linkages in tachyplesin-lipid interactions. Biochemistry. 1993; 32:11704–10.

22). Ramachandran G. Gram-positive and gram-negative bacterial toxins in sepsis: a brief review. Virulence. 2014; 5:213–8.
23). Matsuzaki K. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim Biophys Acta. 1999; 1462:1–10.

24). Wieprecht T, Dathe M, Schümann M, Krause E, Beyermann M, Bienert M. Conformational and functional study of magainin 2 in model membrane environments using the new approach of systematic double-D-amino acid replacement. Biochemistry. 1996; 35:10844–53.

25). Matsuzaki K, Nakamura A, Murase O, Sugishita K, Fujii N, Miyajima K. Modulation of magainin 2-lipid bilayer interactions by peptide charge. Biochemistry. 1997; 36:2104–11.

26). Matsuzaki K, Sugishita K, Ishibe N, Ueha M, Nakata S, Miyajima K, et al. Relationship of membrane curvature to the formation of pores by magainin 2. Biochemistry. 1998; 37:11856–63.

27). Matsuzaki K, Mitani Y, Akada KY, Murase O, Yoneyama S, Zasloff M, et al. Mechanism of synergism between antimicrobial peptides magainin 2 and PGLa. Biochemistry. 1998; 37:15144–53.

28). Matsuzaki K. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim Biophys Acta. 1999; 1462:1–10.

29). Rana FR, Blazyk J. Interactions between the antimicrobial peptide, magainin 2, and Salmonella typhimurium lipopoly-saccharides. FEBS Lett. 1991; 293:11–5.
30). Rana FR, Macias EA, Sultany CM, Modzrakowski MC, Blazyk J. Interactions between magainin 2 and Salmonella typhimurium outer membranes: effect of lipopolysaccharide structure. Biochemistry. 1991; 30:5858–66.

31). Miyata T, Tokunaga F, Yoneya T, Yoshikawa K, Iwanaga S, Niwa M, et al. Antimicrobial peptides, isolated from horseshoe crab hemocytes, tachyplesin II, and polyphemusins I and II: chemical structures and biological activity. J Biochem. 1989; 106:663–8.
32). Matsuzaki K, Sugishita K, Harada M, Fujii N, Miyajima K. Interactions of an antimicrobial peptide, magainin 2, with outer and inner membranes of Gram-negative bacteria. Biochim Biophys Acta. 1997; 1327:119–30.

33). Imura Y, Nishida M, Ogawa Y, Takakura Y, Matsuzaki K. Action mechanism of tachyplesin I and effects of PEGylation. Biochim Biophys Acta. 2007; 1768:1160–9.

34). Imura Y, Nishida M, Matsuzaki K. Action mechanism of PEGylated magainin 2 analogue peptide. Biochim Biophys Acta. 2007; 1768:2578–85.

35). Han E, Lee H. Effects of PEGylation on the binding interaction of magainin 2 and tachyplesin I with lipid bilayer surface. Langmuir. 2013; 29:14214–21.

36). Cruciani RA, Barker JL, Zasloff M, Chen HC, Colamonici O. Antibiotic magainins exert cytolytic activity against transformed cell lines through channel formation. Proc Natl Acad Sci U S A. 1991; 88:3792–6.

37). Lacroix M, Leclercq G. Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat. 2004; 83:249–89.

38). Anghel R, Jitaru D, Bădescu L, Bădescu M, Ciocoiu M. The cytotoxic effect of magainin II on the MDA-MB-231 and M14K tumour cell lines. Biomed Res Int. 2013; 2013:831709.

39). Zanetti M, Gennaro R, Romeo D. Cathelicidins: a novel protein family with a common proregion and a variable C-terminal antimicrobial domain. FEBS Lett. 1995; 374:1–5.

40). Zaiou M, Gallo RL. Cathelicidins, essential gene-encoded mammalian antibiotics. J Mol Med (Berl). 2002; 80:549–61.

41). Gallo RL, Ono M, Povsic T, Page C, Eriksson E, Klagsbrun M, et al. Syndecans, cell surface heparan sulfate proteoglycans, are induced by a proline-rich antimicrobial peptide from wounds. Proc Natl Acad Sci U S A. 1994; 91:11035–9.

42). Frohm M, Agerberth B, Ahangari G, Stâhle-Bäckdahl M, Lidén S, Wigzell H, et al. The expression of the gene coding for the antibacterial peptide LL-37 is induced in human keratinocytes during inflammatory disorders. J Biol Chem. 1997; 272:15258–63.

43). Murakami M, Ohtake T, Dorschner RA, Schittek B, Garbe C, Gallo RL. Cathelicidin anti-microbial peptide expression in sweat, an innate defense system for the skin. J Invest Dermatol. 2002; 119:1090–5.

44). Sørensen OE, Follin P, Johnsen AH, Calafat J, Tjabringa GS, Hiemstra PS, et al. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood. 2001; 97:3951–9.

45). Zaiou M, Nizet V, Gallo RL. Antimicrobial and protease inhibitory functions of the human cathelicidin (hCAP18/LL-37) prosequence. J Invest Dermatol. 2003; 120:810–6.

46). Murakami M, Ohtake T, Dorschner RA, Gallo RL. Cathelicidin antimicrobial peptides are expressed in salivary glands and saliva. J Dent Res. 2002; 81:845–50.

47). Murakami M, Lopez-Garcia B, Braff M, Dorschner RA, Gallo RL. Postsecretory processing generates multiple cathelicidins for enhanced topical antimicrobial defense. J Immunol. 2004; 172:3070–7.

48). Henzler Wildman KA, Lee DK, Ramamoorthy A. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37. Biochemistry. 2003; 42:6545–58.

49). Thennarasu S, Tan A, Penumatchu R, Shelburne CE, Heyl DL, Ramamoorthy A. Antimicrobial and membrane disrupting activities of a peptide derived from the human cathelicidin antimicrobial peptide LL37. Biophys J. 2010; 98:248–57.

50). Murphy PM. The molecular biology of leukocyte chemoattractant receptors. Annu Rev Immunol. 1994; 12:593–633.
51). Zlotnik A, Morales J, Hedrick JA. Recent advances in chemokines and chemokine receptors. Crit Rev Immunol. 1999; 19:1–47.

52). Bals R, Weiner DJ, Meegalla RL, Wilson JM. Transfer of a cathelicidin peptide antibiotic gene restores bacterial killing in a cystic fibrosis xenograft model. J Clin Invest. 1999; 103:1113–7.

53). Frohm Nilsson M, Sandstedt B, Sørensen O, Weber G, Borregaard N, Ståhle-Bäckdahl M. The human cationic antimicrobial protein (hCAP18), a peptide antibiotic, is widely expressed in human squamous epithelia and colocalizes with interleukin-6. Infect Immun. 1999; 67:2561–6.

54). Johansson J, Gudmundsson GH, Rottenberg ME, Berndt KD, Agerberth B. Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J Biol Chem. 1998; 273:3718–24.

55). Oren Z, Lerman JC, Gudmundsson GH, Agerberth B, Shai Y. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity. Biochem J. 1999; 341:501–13.

56). Travis SM, Anderson NN, Forsyth WR, Espiritu C, Conway BD, Greenberg EP, et al. Bactericidal activity of mammalian cathelicidin-derived peptides. Infect Immun. 2000; 68:2748–55.

58). Schumann RR, Leong SR, Flaggs GW, Gray PW, Wright SD, Mathison JC, et al. Structure and function of lipopolysaccharide binding protein. Science. 1990; 249:1429–31.

59). Tobias PS, Ulevitch RJ. Lipopolysaccharide binding protein and CD14 in LPS dependent macrophage activation. Immunobiology. 1993; 187:227–32.

60). Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990; 249:1431–3.

61). Hailman E, Lichenstein HS, Wurfel MM, Miller DS, Johnson DA, Kelley M, et al. Lipopolysaccharide (LPS)-binding protein accelerates the binding of LPS to CD14. J Exp Med. 1994; 179:269–77.

62). Jiang Q, Akashi S, Miyake K, Petty HR. Lipopolysaccharide induces physical proximity between CD14 and toll-like receptor 4 (TLR4) prior to nuclear translocation of NF-kappa B. J Immunol. 2000; 165:3541–4.
63). Lee HK, Dunzendorfer S, Tobias PS. Cytoplasmic domain-mediated dimerizations of toll-like receptor 4 observed by beta-lactamase enzyme fragment complementation. J Biol Chem. 2004; 279:10564–74.
65). Hardaway RM. A review of septic shock. Am Surg. 2000; 66:22–9.
66). Kirikae T, Hirata M, Yamasu H, Kirikae F, Tamura H, Kayama F, et al. Protective effects of a human 18-kilodalton cationic antimicrobial protein (CAP18)-derived peptide against murine endotoxemia. Infect Immun. 1998; 66:1861–8.

67). Nagaoka I, Hirota S, Niyonsaba F, Hirata M, Adachi Y, Tamura H, et al. Cathelicidin family of antibacterial peptides CAP18 and CAP11 inhibit the expression of TNF-alpha by blocking the binding of LPS to CD14 (+) cells. J Immunol. 2001; 167:3329–38.
68). Sawa T, Kurahashi K, Ohara M, Gropper MA, Doshi V, Larrick JW, et al. Evaluation of antimicrobial and lipopolysaccharide-neutralizing effects of synthetic CAP18 fragment against Pseudomonas aeruginosa in a mouse model. Antimcrob Agents Chemother. 1998; 42:3269–75.
69). Lu N, Yang K, Yuan B, Ma Y. Molecular response and cooperative behavior during the interactions of melittin with a membrane: dissipative quartz crystal microbalance experiments and simulations. J Phys Chem B. 2012; 116:9432–8.

70). Habermann E, Reiz KG. On the biochemistry of bee venom peptides, melittin and apamin. Biochem Z. 1965; 343:192–203.
71). Gevod VS, Birdi KS. Melittin and the 8–26 fragment. Differences in ionophoric properties as measured by monolayer method. Biophys J. 1984; 45:1079–83.

72). Sessa G, Freer JH, Colacicco G, Weissmann G. Interaction of alytic polypeptide, melittin, with lipid membrane systems. J Biol Chem. 1969; 244:3575–82.
73). Dathe M, Wieprecht T. Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells. Biochim Biophys Acta. 1999; 1462:71–87.

74). Lauterwein J, Brown LR, W?thrich K. High-resolution 1H-NMR studies of monomeric melittin in aqueous solution. Biochim Biophys Acta. 1980; 622:219–30.

75). Drake AF, Hider RC. The structure of melittin in lipid bilayer membranes. Biochim Biophys Acta. 1979; 555:371–3.

76). Ladokhin AS, White SH. Folding of amphipathic alpha-helices on membranes: energetics of helix formation by melittin. J Mol Biol. 1999; 285:1363–9.
77). Bechinger B. Structure and functions of channel-forming peptides: magainins, cecropins, melittin and alamethicin. J Membr Biol. 1997; 156:197–211.

78). Bechinger B, Lohner K. Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim Biophys Acta. 2006; 1758:1529–39.

79). Killion JJ, Dunn JD. Differential cytolysis of murine spleen, bone-marrow and leukemia cells by melittin reveals differences in membrane topography. Biochem Biophys Res Commun. 1986; 139:222–7.

80). Son DJ, Lee JW, Lee YH, Song HS, Lee CK, Hong JT. Therapeutic application of anti-arthritis, pain-releasing, and anti-cancer effects of bee venom and its constituent compounds. Pharmacol Ther. 2007; 115:246–70.

82). Simmaco M, Mignogna G, Canofeni S, Miele R, Mangoni ML, Barra D. Temporins, antimicrobial peptides from the European red frog Rana temporaria. Eur J Biochem. 1996; 242:788–92.

83). Krishnakumari V, Nagaraj R. Antimicrobial and hemolytic activities of crabrolin, a 13-residue peptide from the venom of the European hornet, Vespa crabro, and its analogs. J Pept Res. 1997; 50:88–93.

84). Argiolas A, Pisano JJ. Isolation and characterization of two new peptides, mastoparan C and crabrolin, from the venom of the European hornet, Vespa crabro. J Biol Chem. 1984; 259:10106–11.

85). Kuchler K, Kreil G, Sures I. The genes for the frog skin peptides GLa, xenopsin, levitide and caerulein contain a homologous export exon encoding a signal sequence and part of an amphiphilic peptide. Eur J Biochem. 1989; 179:281–5.

86). Skerlavaj B, Benincasa M, Risso A, Zanetti M, Gennaro R. SMAP-29: a potent antibacterial and antifungal peptide from sheep leukocytes. FEBS Lett. 1999; 463:58–62.

87). Oren Z, Lerman JC, Gudmundsson GH, Agerberth B, Shai Y. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity. Biochem J. 1999; 341:501–13.

88). Chen Q, Wade D, Kurosaka K, Wang ZY, Oppenheim JJ, Yang D. Temporin A and related frog antimicrobial peptides use formyl peptide receptor-like 1 as a receptor to chemoattract phagocytes. J Immunol. 2004; 173:2652–9.

89). Rosenfeld Y, Shai Y. Lipopolysaccharide (Endotoxin)-host defense antibacterial peptides interactions: role in bacterial resistance and prevention of sepsis. Biochim Biophys Acta. 2006; 1758:1513–22.

90). Trent MS. Biosynthesis, transport, and modification of lipid A. Biochem Cell Biol. 2004; 82:71–86.

91). Rietschel ET, Kirikae T, Schade FU, Mamat U, Schmidt G, Loppnow H, et al. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 1994; 8:217–25.

92). Mangoni ML, Papo N, Barra D, Simmaco M, Bozzi A, Di Giulio A, et al. Effects of the antimicrobial peptide temporin L on cell morphology, membrane permeability and viability of Escherichia coli. Biochem J. 2004; 380:859–65.
93). Weidenmaier C, Kokai-Kun JF, Kulauzovic E, Kohler T, Thumm G, Stoll H, et al. Differential roles of sortaseanchored surface proteins and wall teichoic acid in Staphylococcus aureus nasal colonization. Int J Med Microbiol. 2008; 298:505–13.
94). Neuhaus FC, Baddiley J. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in grampositive bacteria. Microbiol Mol Biol Rev. 2003; 67:686–723.
96). Xia G, Kohler T, Peschel A. The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus. Int J Med Microbiol. 2010; 300:148–54.
97). Su SC, Hua KF, Lee H, Chao LK, Tan SK, Lee H, et al. LTA and LPS mediated activation of protein kinases in the regulation of inflammatory cytokines expression in macrophages. Clin Chim Acta. 2006; 374:106–15.

98). Liljeroos M, Vuolteenaho R, Morath S, Hartung T, Hallman M, Ojaniemi M. Bruton's tyrosine kinase together with PI 3-kinase are part of Toll-like receptor 2 multiprotein complex and mediate LTA induced Toll-like receptor 2 responses in macrophages. Cell Signal. 2007; 19:625–33.

99). Lee IT, Wang SW, Lee CW, Chang CC, Lin CC, Luo SF, et al. Lipoteichoic acid induces HO-1 expression via the TLR2/MyD88/c-Src/NADPH oxidase pathway and Nrf2 in human tracheal smooth muscle cells. J Immunol. 2008; 181:5098–110.

100). Nell MJ, Tjabringa GS, Vonk MJ, Hiemstra PS, Grote JJ. Bacterial products increase expression of the human cathelicidin hCAP-18/LL-37 in cultured human sinus epithelial cells. FEMS Immunol Med Microbiol. 2004; 42:225–31.

101). Agerberth B, Charo J, Werr J, Olsson B, Idali F, Lindbom L, et al. The human antimicrobial and chemotactic peptides LL-37 and alpha-defensins are expressed by specific lymphocyte and monocyte populations. Blood. 2000; 96:3086–93.
Go to : 
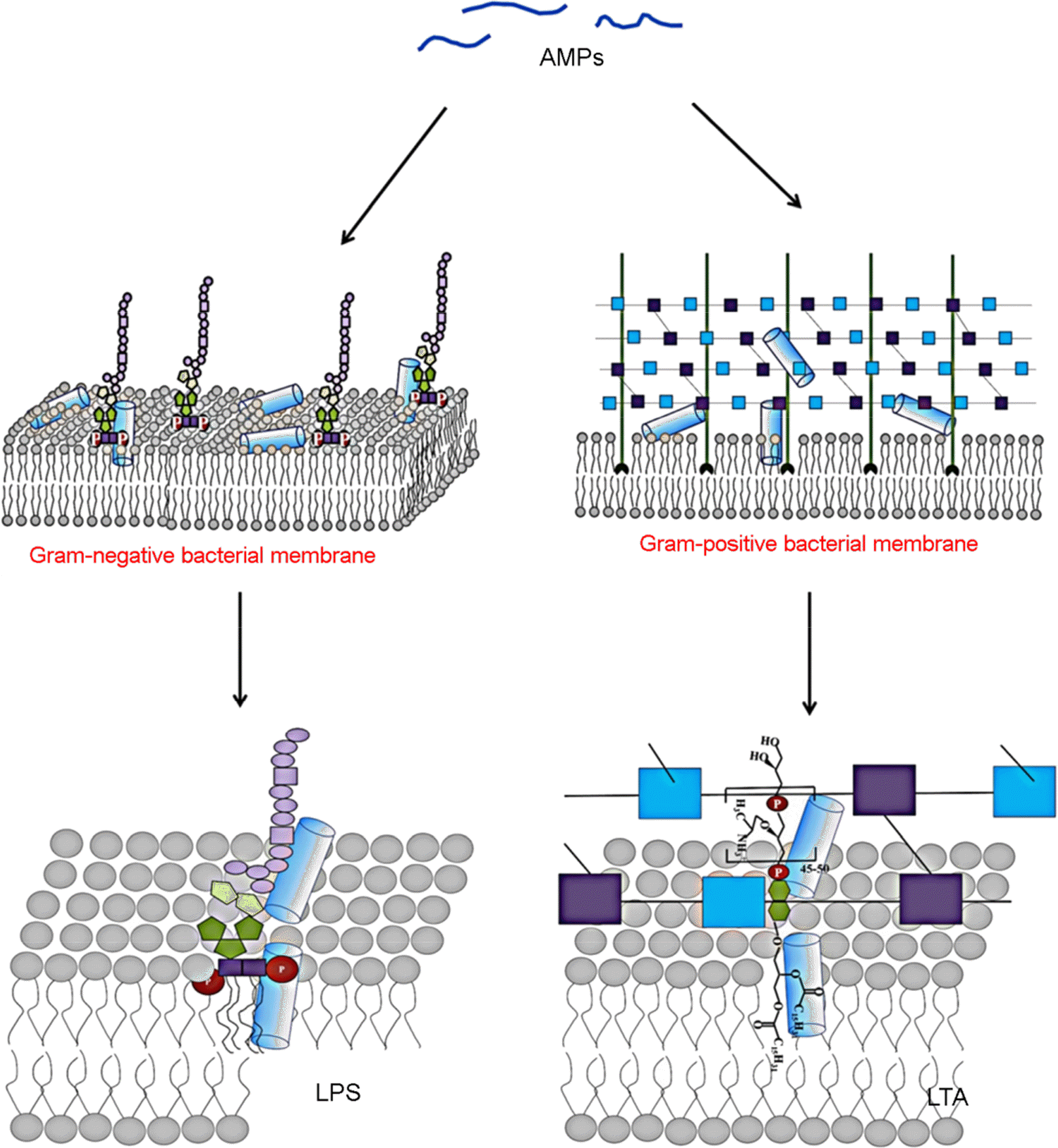 | Figure 2.Binding affinity of antimicrobial peptide on the lipopolysaccharides (LPS) and lipoteichoic acid (LTA) in membranes of Gram negative and Gram-positive bacteria. |
Table 1.
Characterization, activity, and mechanism of antimicrobial peptides.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download