Abstract
Whether we are aware or not, diverse microorganisms are living on almost all environmentally exposed surfaces on our body without eliciting harmful immune responses. In fact, recent understanding from numerous studies indicates that our health is highly dependent on the contribution of intestinal commensal bacteria. It appears through its symbiotic interaction with the host, which is the result of millions of years of co-evolution, the microbiota shapes the immune system. In this review, we discuss the relationship between host physiology and commensal bacteria and explore the molecular mechanisms by which the adaptive immune system is influenced by the intestinal microbiota.
Go to : 
REFERENCES
1). Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010; 7:265–76.

2). Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011; 23:473–80.

3). Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012; 13:260–70.

4). Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002; 347:911–20.

5). Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010; 10:159–69.
6). Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011; 334:255–8.
7). Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004; 303:1662–5.

8). Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013; 13:790–801.

9). Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012; 148:1258–70.

10). Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007; 449:804–10.

11). Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007; 449:811–8.

12). Morgan XC, Segata N, Huttenhower C. Biodiversity and functional genomics in the human microbiome. Trends Genet. 2013; 29:51–8.

13). Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009; 9:313–23.

14). Faith JJ, Rey FE, O'Donnell D, Karlsson M, McNulty NP, Kallstrom G, et al. Creating and characterizing communities of human gut microbes in gnotobiotic mice. ISME J. 2010; 4:1094–8.

15). Wostmann BS, Larkin C, Moriarty A, Bruckner-Kardoss E. Dietary intake, energy metabolism, and excretory losses of adult male germfree Wistar rats. Lab Anim Sci. 1983; 33:46–50.
16). Diaz Heijtz R, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A. 2011; 108:3047–52.
17). Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010; 330:1768–73.

18). Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007; 2:119–29.

19). Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, et al. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011; 60:631–7.

20). Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011; 145:745–57.

21). Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007; 131:33–45.

22). Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012; 487:104–8.

23). Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006; 444:1027–31.

24). Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006; 444:1022–3.
25). Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010; 32:815–27.

26). Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011; 108(Suppl 1):4615–22.

27). Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010; 107:12204–9.

28). Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008; 453:620–5.

29). Ochoa-Repáraz J, Mielcarz DW, Ditrio LE, Burroughs AR, Begum-Haque S, Dasgupta S, et al. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol. 2010; 185:4101–8.
30). Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011; 332:974–7.

31). Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011; 331:337–41.
32). Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013; 500:232–6.
33). Mortensen PB, Clausen MR. Short-chain fatty acids in the human colon: relation to gastrointestinal health and disease. Scand J Gastroenterol Suppl. 1996; 216:132–48.

34). Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013; 341:569–73.
35). Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013; 504:446–50.

36). Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013; 504:451–5.

37). Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014.

38). Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008; 455:808–12.

39). Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, et al. The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe. 2011; 10:260–72.

40). Prakash T, Oshima K, Morita H, Fukuda S, Imaoka A, Kumar N, et al. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of th17 cell differentiation. Cell Host Microbe. 2011; 10:273–84.

41). Blumershine RV, Savage DC. Filamentous microbes indigenous to the murine small bowel: A scanning electron microscopic study of their morphology and attachment to the epithelium. Microb Ecol. 1977; 4:95–103.

42). Koopman JP, Stadhouders AM, Kennis HM, De Boer H. The attachment of filamentous segmented microorganisms to the distal ileum wall of the mouse: a scanning and transmission electron microscopy study. Lab Anim. 1987; 21:48–52.

43). Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol. 2010; 10:735–44.

44). Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009; 31:677–89.

45). Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009; 139:485–98.

46). Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008; 4:337–49.

47). Stepankova R, Powrie F, Kofronova O, Kozakova H, Hudcovic T, Hrncir T, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm Bowel Dis. 2007; 13:1202–11.

48). Jeon SG, Kayama H, Ueda Y, Takahashi T, Asahara T, Tsuji H, et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS pathog. 2012; 8:e1002714.
49). Di Giacinto C, Marinaro M, Sanchez M, Strober W, Boirivant M. Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGF-beta-bearing regulatory cells. J Immunol. 2005; 174:3237–46.
50). Lyons A, O'Mahony D, O'Brien F, MacSharry J, Sheil B, Ceddia M, et al. Bacterial strain-specific induction of Foxp3+ T regulatory cells is protective in murine allergy models. Clin Exp Allergy. 2010; 40:811–9.
Go to : 
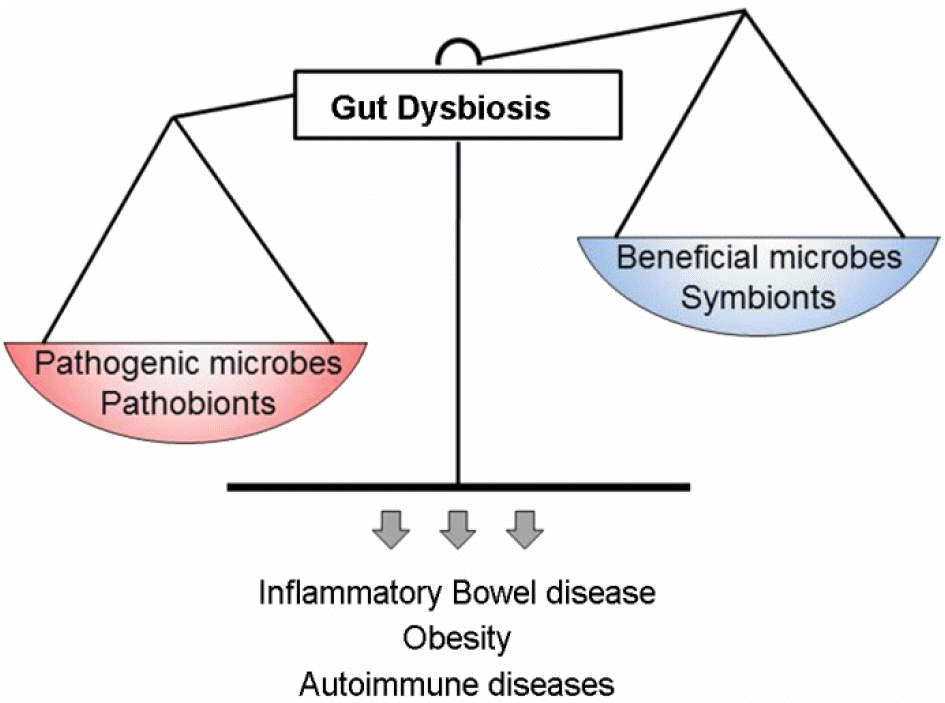 | Figure 1.Gut dysbiosis are associated with human diseases. The microbiota in the mammal gastrointestinal tract can divide to pathogenic and beneficial bacteria called pathobionts and symbionts respectively. A healthy microbiota contains a balanced composition of pathobionts and symbionts. If there is an unnatural shift in the composition of the gut microbiota, gut dysbiosis, can lead to intestinal inflammation, many other diseases and autoimmune diseases. |
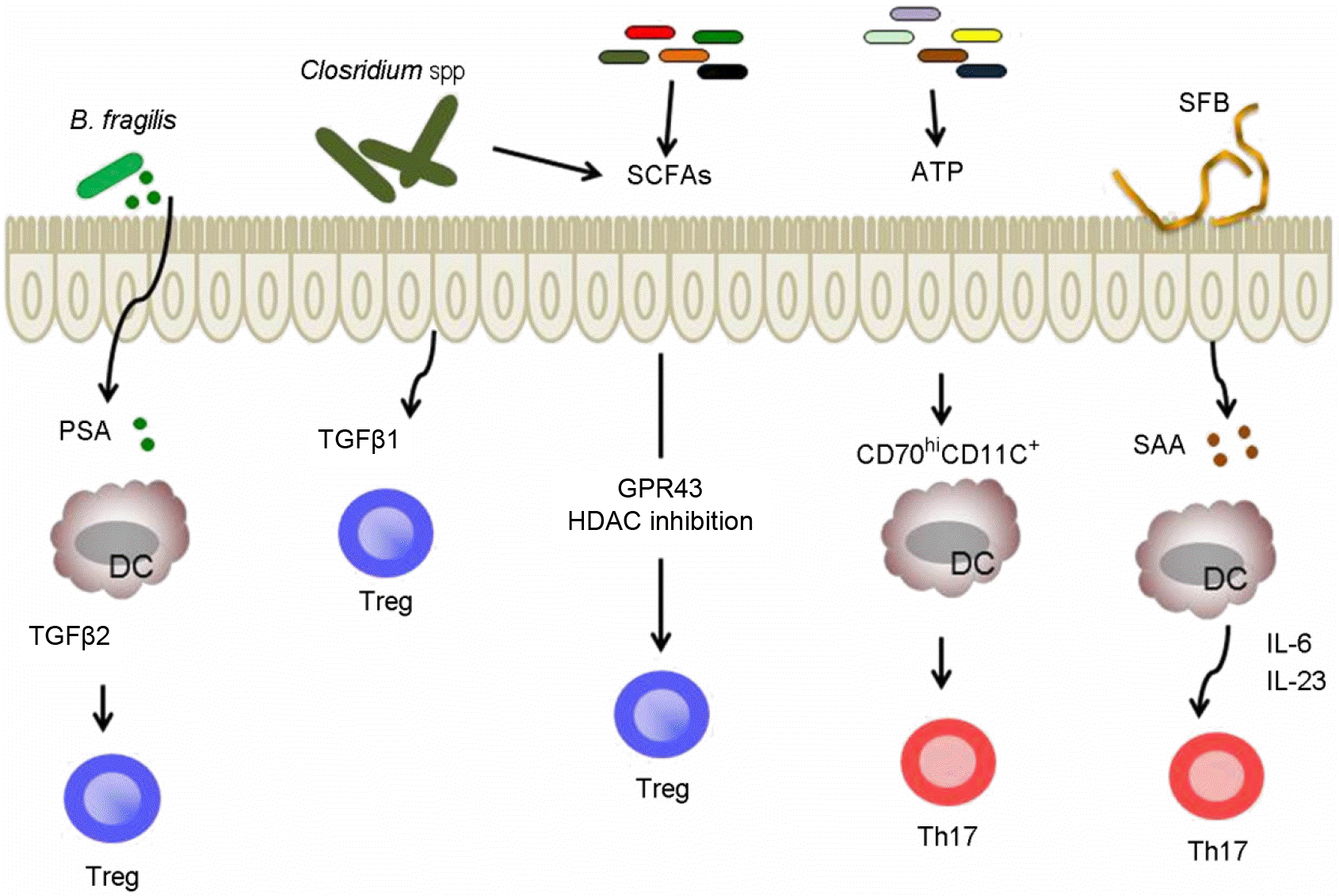 | Figure 2.Immunomodulatory members of the gut microbiota and microbial metabolites regulate the function of CD4+ T cell subsets. B. fragilis or Clostridium spp. promotes the induction of Tregs in colonic laminar propria via PSA-TLR2 signaling or the secretion of TGFβ1 respectively. SCFAs mediate the proliferation and induction of Treg cells in colon through distinct mechanism; GPR43 or HDAC inhibitory activity. Commensal bacteria derived-extracellular ATP induces the differentiation of Th17 cells in colon. Colonization of SFBs promotes the development of Th17 cells in laminar propria of small intestine by inducing the production of SAA. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download