Abstract
REFERENCES
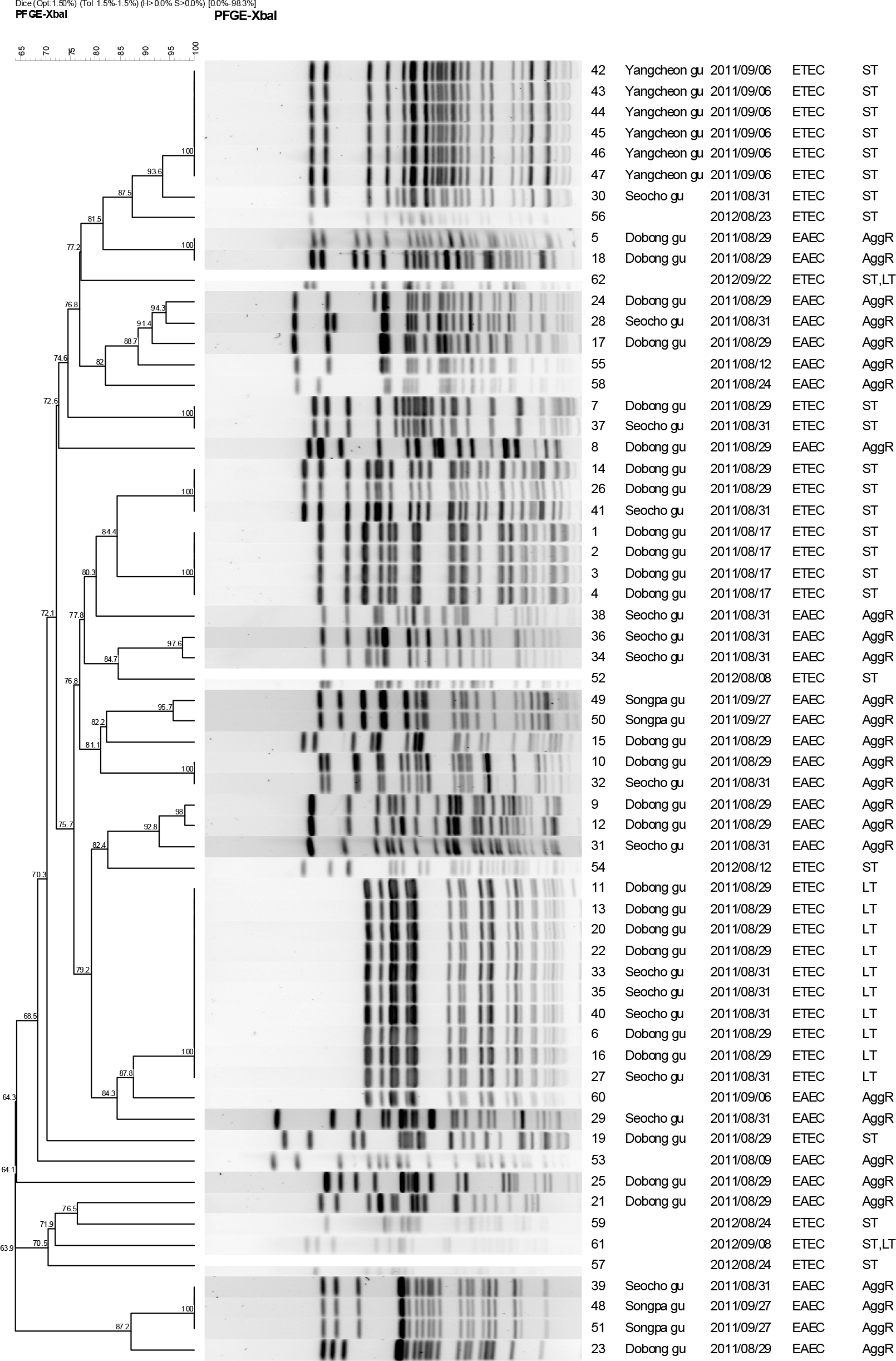 | Figure 1.Dendrogram showing the clustering of PFGE patterns for the 62 ETEC and EAEC isolates. A group indicates 81.5% similarity; B group indicates 82% similarity; C group indicates 80.3% similarity; D group indicates 84.7% similarity; E groups indicate 81.1% similarity; F groups indicate 82.4% similarity; G group indicates 84.3% similarity; H group indicates 87.2% similarity. |
Table 1.
| Case | Group | Region (Gu) | Occurrence place | Receipt date | Pathogenic gene | Selection numbera |
|---|---|---|---|---|---|---|
| Outbreak | O1 | Dobong | Police office | 2011-8-17 |
ST ST |
4/21 4/12 |
| O2 | Dobong | School | 2011-8-29 |
LT aggR |
6/9 12/42 |
|
| O3 | Seocho | School | 2011-8-29 |
ST LT aggR |
3/10 4/6 8/31 |
|
| O4 | Yangcheon | Company | 2011-9-6 | ST | 6/29 | |
| O5 | Songpa | School | 2011-9-27 | aggR | 4/4 | |
| Sporadic | S1 | Gwangjin | Southeast Asia travel | 2011-8-8 | ST | 1 |
| S2 | Gwanak | Southeast Asia travel (Cambodia) | 2011-8-9 | aggR | 1 | |
| S3 | Southeast Asia travel | 2011-8-12 | ST | 1 | ||
| S4 | Yangcheon | Southeast Asia travel | 2011-8-12 | aggR | 1 | |
| S5 | Nowon | Southeast Asia travel (China) | 2011-8-23 | ST | 1 | |
| S6 | Other | 2011-8-24 | ST | 1 | ||
| S7 | Dongjak | Other | 2011-8-24 | aggR | 1 | |
| S8 | Gangdong | Restaurant | 2011-8-24 | ST | 1 | |
| S9 | Seongbuk | Southeast Asia travel (Philippine) | 2011-9-6 | aggR | 1 | |
| S10 | Nowon | Southeast Asia travel | 2011-9-8 | ST, LT | 1 | |
| S11 | Jungnang | Southeast Asia travel | 2011-9-22 | ST, LT | 1 | |
| Total | 62 |
Table 2.
| No.(%) of isolates resistant toa | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AM | AN | SAM | CF | CZ | FEP | CTT | CTX | CIP | C | GM | IPM | NA | TE | TIC | SXT | |
| EAEC (n=28) | 15 (54) | 0 | 0 | 2 (7) | 2 (7) | 0 | 0 | 1 (4) | 0 | 4 (14) | 2 (7) | 1 (4) | 13 (46) | 16 (57) | 15 (54) | 12 (43) |
| ETEC (n=34) | 6 (18) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (3) | 0 | 0 | 16 (47) | 11 (32) | 6 (18) | 5 (15) |
| Total (n=62) | 21 (34) | 0 | 0 | 2 (3.2) | 2 (3.2) | 0 | 0 | 1 (1.6) | 0 | 5 (8.1) | 2 (3.2) | 1 (1.6) | 29 (47) | 27 (44) | 21 (34) | 17 (27) |
a Abbreviations: AM, ampicillin; AN, amikacin; SAM, ampicillin/sulbatam; CF, cephalothin; CZ, cefazolin; FEP, cefepime; CTT, cefotetan; CTX, cefotaxime; CIP, ciprofloxacin; C, chloramphenicol; GM, gentamicin; IPM, imipenem; NA, nalidixic acid; Te, tetracycline; TIC, ticarcillin; SXT, trimethoprim/sulfamethoxazole
Table 3.
| Case | Group | Region (Gu) | Pathogenic gene | Selection number | Antimicrobial resistant patterns (numbers)a |
|---|---|---|---|---|---|
| Outbreak | O1 | Dobong |
ST ST |
4 4 |
TE (4) NA, NA-TE, AM-NA-TE-TIC-SXT |
| O2 | Dobong |
LT aggR ST |
6 12 3 |
− TE, NA-TE, AM-NA-TIC, AM-NA-TIC-SXT, AM-TE-TIC-SXT (2), AM-C-NA-TIC-SXT, AM-NA-TE-TIC-SXT, AM-CF-CZ-C-GM-NA-TIC NA (3) |
|
| O3 | Seocho |
LT aggR |
4 8 |
− TE (2), AM-TIC, NA-TE, NA-TE-SXT, AM-IPM-NA-TE-TIC, AM-GM-TE-TIC-SXT |
|
| O4 | Yangcheon | ST | 6 | NA (5), NA-TE | |
| O5 | Songpa | aggR | 4 | NA-TE-SXT (2), AM-TE-TIC-SXT (2) | |
| Sporadic | S1 | Gwangjinb | ST | 1 | AM-NA-TE-TIC-SXT |
| S2 | Gwanakb | aggR | 1 | AM-CF-CZ-CTX-TE-SXT | |
| S3 | Yangcheonb | ST | 1 | AM-NA-TE-TIC-SXT | |
| S4 | aggR | 1 | AM-C-NA-TIC | ||
| S5 | Nowonb | ST | 1 | NA | |
| S6 | Dongjak | ST | 1 | AM-TE-TIC-SXT | |
| S7 | aggR | 1 | AM-C-NA-TIC-SXT | ||
| S8 | Gangdong | ST | 1 | NA | |
| S9 | Seongbukb | aggR | 1 | − | |
| S10 | Nowonb | ST, LT | 1 | AM-TE-TIC | |
| S11 | Jungnangb | ST, LT | 1 | AM-TIC-SXT |
a Abbreviations: AM, ampicillin; AN, amikacin; SAM, ampicillin/sulbatam; CF, cephalothin; CZ, cefazolin, FEP, cefepime; CTT, cefotetan; CTX, cefotaxime; CIP, ciprofloxacin; C, chloramphenicol; GM, gentamicin; IPM, imipenem; NA, nalidixic acid; Te, tetracycline; TIC, ticarcillin; SXT, trimethoprim/sulfamethoxazole




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download