Abstract
Norovirus and rotavirus are important causes of acute nonbacterial gastroenteritis in communities worldwide. Genotypes of norovirus and rotavirus in stool samples, which were collected from patients, was determined by RT-PCR and ELISA. A total 4,484 stool samples (461 outbreak cases) collected during January 2010-April 2013 were analyzed. Four hundred thirty eight of samples were positive for norovirus. Twelve samples were positive for Rotavirus. Norovirus genotypes identified were nine kinds of G I genotype (G I-1, G I-2, G I-3, G I-4, G I-6, G I-7, G I-9, G I-12, G I-13) and nine kinds of G II genotype(G II-2, G II-4, G II-5, G II-6, G II-7, G II-11, G II-12, G II-15, G II-16). Rotavirus genotypes were G2P[4]. The results might be useful information for evaluating the epidemiological status of viral diarrhea in Seoul, and providing a strategy to prevent human norovirus, rotavirus and other threats to public health in Korea.
Go to : 
REFERENCES
1). Patel MM, Hall AJ, Vinjé J, Parashar UD. Noroviruses: a comprehensive review. J Clin Virol. 2009; 44:1–8.

2). Belliot G, Noel JS, Li JF, Seto Y, Humphrey CD, Ando T, et al. Characterization of capsid genes, expressed in the baculovirus system, of three new genetically distinct strains of “Norwalk-like viruses”. J Clin Microbiol. 2001; 39:4288–95.

3). Wang QH, Han MG, Cheetham S, Souza M, Funk JA, Saif LJ. Porcine noroviruses related to human noroviruses. Emerg Infect Dis. 2005; 11:1874–81.

4). Bull RA, Eden JS, Rawlinson WD, White PA. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 2010; 26(6):e1000831.

5). Abe M, Ito N, Morikawa S, Takasu M, Murase T, Kawashima T, et al. Molecular epidemiology of rotaviruses among healthy calves in Japan: isolation of a novel bovine rotavirus bearing new P and G genotypes. Virus Res. 2009; 144:250–7.

6). Collins PJ, Martella V, Buonavoglia C, O'Shea H. Identification of a G2-like porcine rotavirus bearing a novel VP4 type, P[32]. Vet Res. 2010; 41:73.

7). Ursu K, Kisfali P, Rigó D, Ivanics E, Erdélyi K, Dán A, et al. Molecular analysis of the VP7 gene of pheasant rotaviruses identifies a new genotype, designated G23. Arch Virol. 2009; 154:1365–9.

8). Parashar UD, Gibson CJ, Bresee JS, Glass RI. Rotavirus and severe childhood diarrhea. Emerg Infect Dis. 2006; 12:304–6.

9). Kim SH, Cheon DS, Kim JH, Lee DH, Jheong WH, Heo YJ, et al. Outbreaks of gastroenteritis that occurred during school excursions in Korea were associated with several waterborne strains of Norovirus. J Clin Microbiol. 2005; 43:4836–9.

10). Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, et al. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992; 30:1365–73.

11). Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, et al. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990; 28:276–82.

12). Gong YW, Oh BY, Kim HY, Lee MY, Kim YH, Go JM, et al. Molecular epidemiologic investigation of norovirus infections in Incheon City, Korea, from 2005 to 2007. J Bacteriol Virol. 2008; 38:249–57.

13). Kim NH, Park EH, Park YK, Min SK, Jin SH, Park SH. Study on norovirus genotypes in Busan. Journal of Life Science. 2011; 21:845–50.
14). Park DJ, Kim JS, Park JY, Kim HS, Song W, Kim HS, et al. Epidemiological analysis of norovirus infection between march 2007 and February 2010. Korean J Lab Med. 2010; 30:647–53.

15). Mathijs E, Denayer S, Palmeira L, Botteldoorn N, Scipioni A, Vanderplasschen A, et al. Novel norovirus recombinants and of GII.4 sub-lineages associated with outbreaks between 2006 and 2010 in Belgium. Virol J. 2011; 8:310.

16). Ramirez S, Giammanco GM, De Grazia S, Colomba C, Martella V, Arista S. Emerging GII.4 norovirus variants affect children with diarrhea in Palermo, Italy in 2006. J Med Virol. 2009; 81:139–45.

17). Okame M, Akihara S, Hansman G, Hainian Y, Tran HT, Phan TG, et al. Existence of multiple genotypes associated with acute gastroenteritis during 6-year survey of norovirus infection in Japan. J Med Virol. 2006; 78:1318–24.

18). Chen SY, Chang YC, Lee YS, Chao HC, Tsao KC, Lin TY, et al. Molecular epidemiology and clinical manifestations of viral gastroenteritis in hospitalized pediatric patients in Northern Taiwan. J Clin Microbiol. 2007; 45:2054–7.

19). Jee YM. Norovirus food poisoning and laboratory surveillence for viral gastroenteritis in Korea. Health and Welfare Policy Forum. 2006; 118:26–34.
20). Mounts AW, Ando T, Koopmans M, Bresee JS, Noel J, Glass RI. Cold Weather Seasonality of Gastroenteritis Associated with Norwalk-like Viruses. J Infect Dis. 2000; 181:S284–7.

21). Papaventsis DC, Dove W, Cunliffe NA, Nakagomi O, Combe P, Grosjean P, et al. norovirus infection in children with acute gastroenteritis, Madagascar, 2004–2005. Emerg Infection Dis. 2007; 13:908–11.

22). Ho EC, Cheng PK, Lau AW, Wong AH, Lim WW. Atypical norovirus epidemic in Hong Kong during summer of 2006 caused by a new genogroup II/4 variant. J Clin Microbiol. 2007; 45:2205–11.

23). Pang XL, Preiksaitis JK, Wong S, Li V, Lee BE. Influence of novel norovirus GII.4 variants on gastroenteritis outbreak dynamics in Alberta and the Northern territories, Canada between 2000 and 2008. PLoS One. 2010; 5:e11599.

24). Donia D, Cenko F, Divizia M. Molecular characterization of norovirus GII strains identified in Albania. J Med Virol. 2013; 85:731–6.

25). Bennett S, MacLean A, Miller RS, Aitken C, Gunson RN. Increased norovirus activity in Scotland in 2012 is associated with the emergence of a new norovirus GII.4 variant. Euro Surveill. 2013; 10:18. pii: 20349.

26). Hall AJ, Eisenbart VG, Etingüe AL, Gould LH, Lopman BA, Parashar UD. Epidemiology of foodborne norovirus outbreaks, United States, 2001–2008. Emerg Infect Dis. 2012; 18:1566–73.

27). Van Beek J, Ambert-Balay K, Botteldoorn N, Eden JS, Fonager J, Hewitt J, et al. Indications for worldwide increased norovirus activity associated with emergence of a new variant of genotype II.4, late 2012. Euro Surveill. 2013; 18:8–9.

28). Yu JH, Kim NY, Koh YJ, Lee HJ. Epidemiology of foodborne norovirus outbreak in Incheon, Korea. J Korean Med Sci. 2010; 25:1128–33.

29). White PA, Eden JS, Hansman GS. Molecular epidemiology of noroviruses and sapoviruses and their role in Australian outbreaks of acute gastroenteritis. Microbiology Australia. 2012; 33:70–3.

30). Bull RA, Eden JS, Rawlinson WD, White PA. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 2010; 6:e1000831.

31). Inoue Y, Imanishi Y, Kitahori Y. An outbreak of group A rotavirus G1P[8] in an elementary school, Nara prefecture. Jpn J Infect Dis. 2008; 61:426.
32). Lee JM, Kim HY, Lee MY, Lee KB, Cheon DS, Jee YM. The prevalence and genotypic distribution of group A rotavirus detected from patients with acute gastroenteritis patients in Incheon. J Bacteriol Virol. 2007; 37:39–45.

33). Choi HJ, Oh BY, Lee MY, Koh YJ, Gong YW, Hur MJ, et al. The prevalence and distribution of the P and G genotypes of a group A rotavirus detected in acute gastroenteritis patients from Incheon. Journal of Life Science. 2012; 22:600–4.

34). Han TH, Kim CH, Chung JY, Park SH, Hwang ES. Genetic characterization of rotavirus in children in South Korea from 2007 to 2009. Arch Virol. 2010; 155:1663–73.

35). Kawai K, O'Brien MA, Goveia MG, Mast TC, El Khoury AC. Burden of rotavirus gastroenteritis and distribution of rotavirus strains in Asia: A systematic review. Vaccine. 2012; 30:1244–54.

36). Cardemil CV, Cortese MM, Medina-Marino A, Jasuja S, Desai R, Leung J, et al. Two rotavirus outbreaks caused by genotype G2P[4] at large retirement communities: cohort studies. Ann Intern Med. 2012; 157:621–31.
37). Yan H, Abe T, Phan TG, Nguyen TA, Iso T, Ikezawa Y, et al. Outbreak of acute gastroenteritis associated with group A rotavirus and genogroup I sapovirus among adults in a mental health care facility in Japan. J Med Virol. 2005; 75:475–81.

38). Marshall J, Botes J, Gorrie G, Boardman C, Gregory J, Griffith J, et al. Rotavirus detection and characterisation in outbreaks of gastroenteritis in aged-care facilities. J Clin Virol. 2003; 28:331–40.

Go to : 
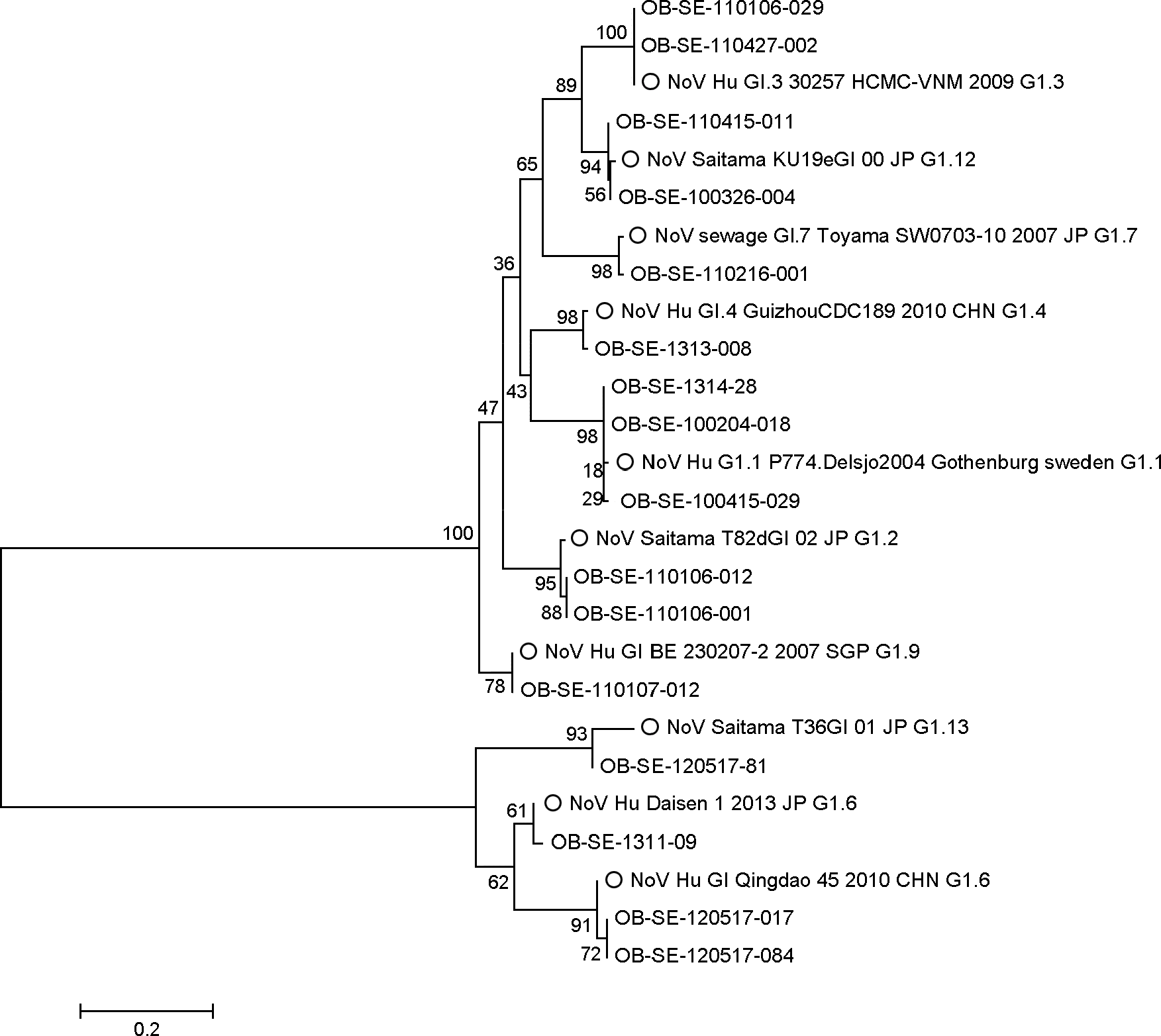 | Figure 3.Phylogenetic analysis of Norovirus G I strains. Accession number of reference strains from Genbank (EU085529, AB112131, HE716741, JX074065, JQ359761, AB779748, AB504701, GQ925127, AB058529, and AB112133). OB means outbreak, SE means Seoul province. Reference strains are indicated by the circles. |
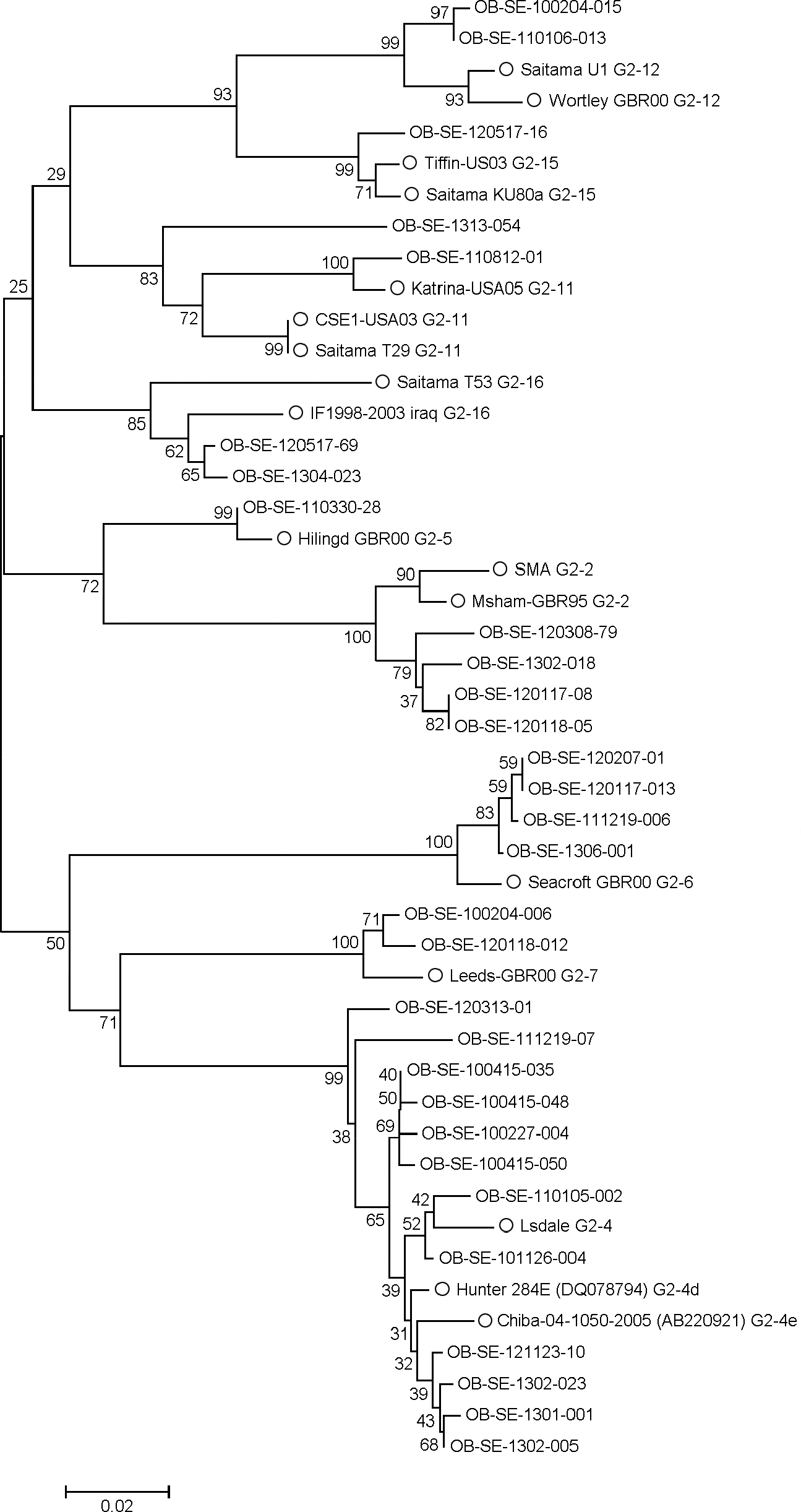 | Figure 4.Phylogenetic analysis of Norovirus G II strains. Accession number of reference strains from Genbank (U70059, X81879, AB220921, DQ078794, X86557, AJ277607, AJ277620, AJ277608, DQ438972, AY502009, AB112221, AB039775, AJ277618, AY502010, AB058582, AY675554, and AB112260). OB means outbreak, SE means Seoul province. Reference strains are indicated by the circles. |
Table 1.
Primers used for the detection of Norovirus
Table 2.
Primers used for the detection of Rotavirus
Table 3.
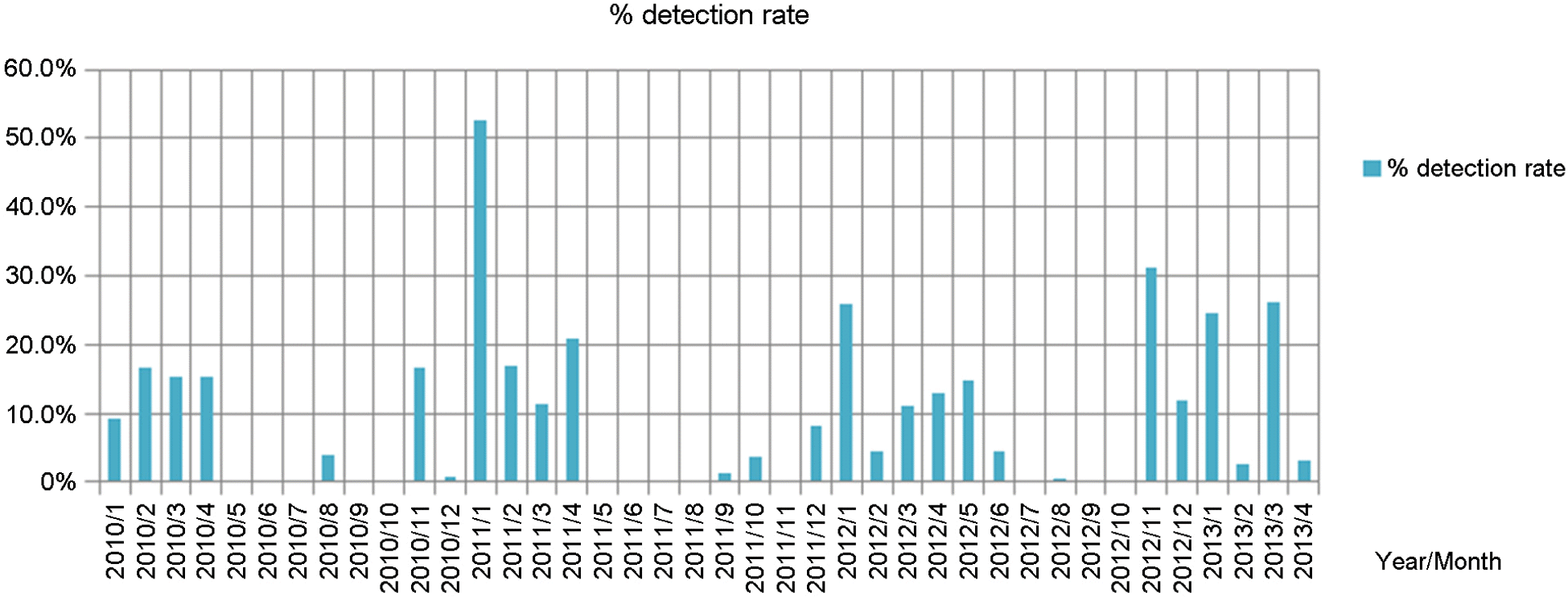
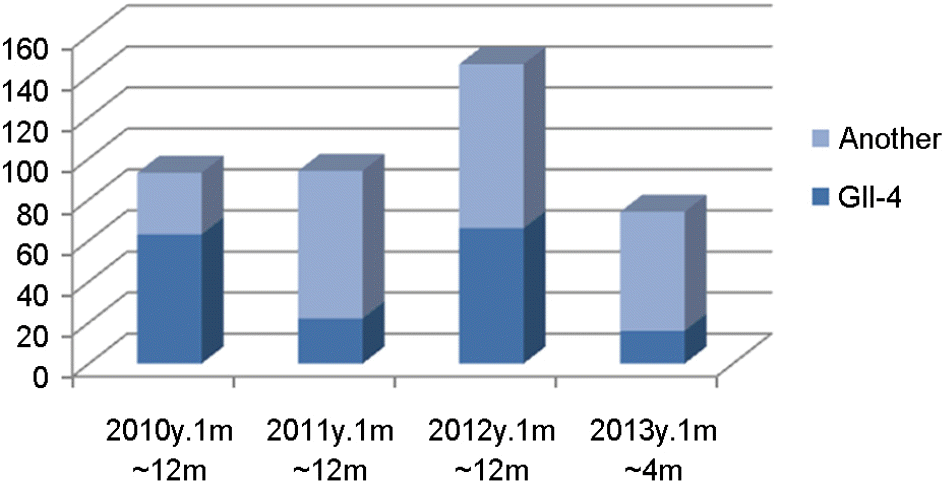
The number of outbreak cases and Norovirus positive cases including genotype
| Year | Month | Case tested | Positive cases | Genotype |
|---|---|---|---|---|
| 1 | 3 | 1 | GII-NTa | |
| 2 | 7 | 3 | GI-1, GI-6, GII-NT, GII-4, GII-7, GII-12 | |
| 3 | 7 | 1 | GI-12 | |
| 4 | 13 | 1 | GII-4 | |
| 5 | 5 | 0 | ||
| 2010 |
6 7 |
12 26 |
0 0 |
|
| 8 | 22 | 2 | GI-12, GII-NT | |
| 9 | 17 | 0 | ||
| 10 | 12 | 0 | ||
| 11 | 9 | 1 | GII-NT, GII-4 | |
| 12 | 15 | 1 | GI-NT, GII-NT | |
| Subtotal | 148 | 10 | ||
| 1 | 7 | 6 | GI-1, GI-2, GI-3, GI-9, GII-NT, GII-2, GII-4, GII-5, GII-12 | |
| 2 | 10 | 4 | GI-7, GI-NT, GII-7 | |
| 3 | 16 | 2 | GII-2, GII-5 | |
| 4 | 2 | 2 | GI-1, GI-3, GI-4, GI-12, GII-5 | |
| 2011 |
5 6 |
7 11 |
0 0 |
|
| 7 | 12 | 0 | ||
| 8 | 21 | 1 | GI-3, GII-11 | |
| 9 | 11 | 1 | GII-4 | |
| 10 | 9 | 1 | GII-2 | |
| 11 | 3 | 0 | ||
| 12 | 12 | 2 | GII-6 | |
| Subtotal | 121 | 19 | ||
| 1 | 12 | 3 | GI-6, GII-2, GII-4, GII-6 | |
| 2 | 14 | 3 | GII-NT, GII-4, GII-6 | |
| 3 | 7 | 3 | GI-3, GII-2, GII-4 | |
| 2012 | 4 | 6 | 2 | GI-7, GI-12, GII-NT |
| 5 | 12 | 2 | GI-6, GI-13, GII-NT, GII-15, GII-16 | |
| 6 | 9 | 1 | GII-2 | |
| 7 | 5 | 0 | ||
| 8 | 14 | 1 | GII-4 | |
| 9 | 16 | 0 | ||
| 2012 | 10 | 17 | 0 | |
| 11 | 11 | 2 | GII-4 | |
| 12 | 17 | 6 | GII-4, GII-11 | |
| Subtotal | 140 | 23 | ||
| GI-NT, GII-NT, GII-2, GII-4, GII-6, GII-11, GII-16 | ||||
| 1 | 22 | 10 | ||
| 2013 | 2 | 11 | 1 | GI-NT |
| 3 | 7 | 1 | GI-4, GII-11, GII-16 | |
| 4 | 12 | 1 | GI-1, GII-4 | |
| Subtotal | 52 | 13 | ||
| Total | 461 | 65 | ||




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download