Abstract
Although viruses are the most common cause of acute gastroenteritis (AGE) in humans, details about the causative viruses in AGE are largely unknown because many causative viruses are unable to be cultured by current culture techniques. In our study, fecal samples from 10 children under five years of age with unexplained AGE and 10 healthy children were investigated for RNA viruses using random priming (RP)-mediated sequence-independent single primer amplification (SISPA). The causative viruses in cases of cryptogenic diarrhea were then assessed for their potential diagnostic value. Of the 1,129 viral clones identified, rotavirus was most commonly associated with AGE (125 sequences, 22.4%). In contrast, bacteriophage was most common (43 sequences, 13.6%) in healthy children. The remaining 515 viral clones were unidentifiable. These findings suggest that investigation of cases or outbreaks of unexplained diarrhea using a metaviromic strategy is a new avenue for diagnosis.
Go to : 
REFERENCES
1). Chang KO, George DW, Patton JB, Green KY, Sosnovtsev SV. Leader of the capsid protein in feline calicivirus promotes replication of Norwalk virus in cell culture. J Virol. 2008; 82:9306–17.

2). Shim JO, Baek IH, Le VP, Ko EM, Seok WS, Uh Y, et al. Molecular characterization of rotavirus diarrhea among children in South Korea: detection of an unusual G11 strain. Arch Virol. 2011; 156:887–92.

3). Zhang T, Breitbart M, Lee WH, Run JQ, Wei CL, Soh SW, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006; 4:e3.

4). Kim W. Application of metagenomic techniques: understanding the unrevealed human microbiota and explaining in the clinical infectious diseases. J Bacteriol Virol. 2012; 42:263–75.
5). Bench SR, Hanson TE, Williamson KE, Ghosh D, Radosovich M, Wang K, et al. Metagenomic characterization of Chesapeake Bay virioplankton. Appl Environ Microbiol. 2007; 73:7629–41.

6). Day JM, Ballard LL, Duke MV, Scheffler BE, Zsak L. Metagenomic analysis of the turkey gut RNA virus community. Virol J. 2010; 7:313.

7). Ng TF, Willner DL, Lim YW, Schmieder R, Chau B, Nilsson C, et al. Broad Surveys of DNA Viral Diversity Obtained through Viral Metagenomics of Mosquitoes. PLoS One. 2011; 6:e20579.

8). Park EJ, Kim KH, Abell GC, Kim MS, Roh SW, Bae JW. Metagenomic analysis of the viral communities in fermented foods. Appl Environ Microbiol. 2011; 77:1284–91.

9). Greninger AL, Chen EC, Sittler T, Scheinerman A, Roubinian N, Yu G, et al. A metagenomic analysis of pandemic influenza A (2009 H1N1) infection in patients from North America. PLoS One. 2010; 5:e13381.

10). Ramani S, Kang G. Viruses causing childhood diarrhoea in the developing world. Curr Opin Infect Dis. 2009; 22:477–82.

11). Lee JI, Park SH, Kim MS, Oh YH, Yu IS, Choi BH, et al. Surveillance of Acute Gastroenteritis in Seoul, Korea, During May 2004 and June 2007. J Bacteriol Virol. 2009; 39:363–71.

12). Djikeng A, Halpin R, Kuzmickas R, Depasse J, Feldblyum J, Sengamalay N, et al. Viral genome sequencing by random priming methods. BMC Genomics. 2008; 9:5.

13). Wei CJ, Boyington JC, Dai K, Houser KV, Pearce MB, Kong WP, et al. Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci Transl Med. 2010; 2:24ra1.

14). Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005; 3:431–8.

16). Streit WR, Schmitz RA. Metagenomics–the key to the uncultured microbes. Curr Opin Microbiol. 2004; 7:492–8.
17). Kittigul L, Pombubpa K, Taweekate Y, Diraphat P, Sujirarat D, Khamrin P, et al. Norovirus GII-4 2006b variant circulating in patients with acute gastroenteritis in Thailand during a 2006–2007 study. J Med Virol. 2010; 82:854–60.

18). Kittigul L, Ekchaloemkiet S, Utrarachkij F, Siripanichgon K, Sujirarat D, Pungchitton S, et al. An efficient virus concentration method and RT-nested PCR for detection of rotaviruses in environmental water samples. J Virol Methods. 2005; 124:117–22.

19). Harada S, Okada M, Yahiro S, Nishimura K, Matsuo S, Miyasaka J, et al. Surveillance of pathogens in outpatients with gastroenteritis and characterization of sapovirus strains between 2002 and 2007 in Kumamoto Prefecture, Japan. J Med Virol. 2009; 81:1117–27.

20). r-biopharm. RIDASCREEN Rotavirus Enzyme immunoassay for the detection of Rotavirus. R-Biopharm AG, Darmstadt. 2004; C 0901.
21). Chinsangaram J, Akita GY, Castro AE, Osburn BI. PCR detection of group A bovine rotaviruses in feces. J Vet Diagn Invest. 1993; 5:516–21.

22). Nix WA, Maher K, Pallansch MA, Oberste MS. Parechovirus typing in clinical specimens by nested or semi-nested PCR coupled with sequencing. J Clin Virol. 2010; 48:202–7.

23). Li L, Victoria J, Kapoor A, Naeem A, Shaukat S, Sharif S, et al. Genomic characterization of novel human parechovirus type. Emerg Infect Dis. 2009; 15:288–91.

24). Shu Y, Li CK, Li Z, Gao R, Liang Q, Zhang Y, et al. Avian influenza A(H5N1) viruses can directly infect and replicate in human gut tissues. J Infect Dis. 2010; 201:1173–7.

25). Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, et al. Viral diversity and dynamics in an infant gut. Res Microbiol. 2008; 159:367–73.

Go to : 
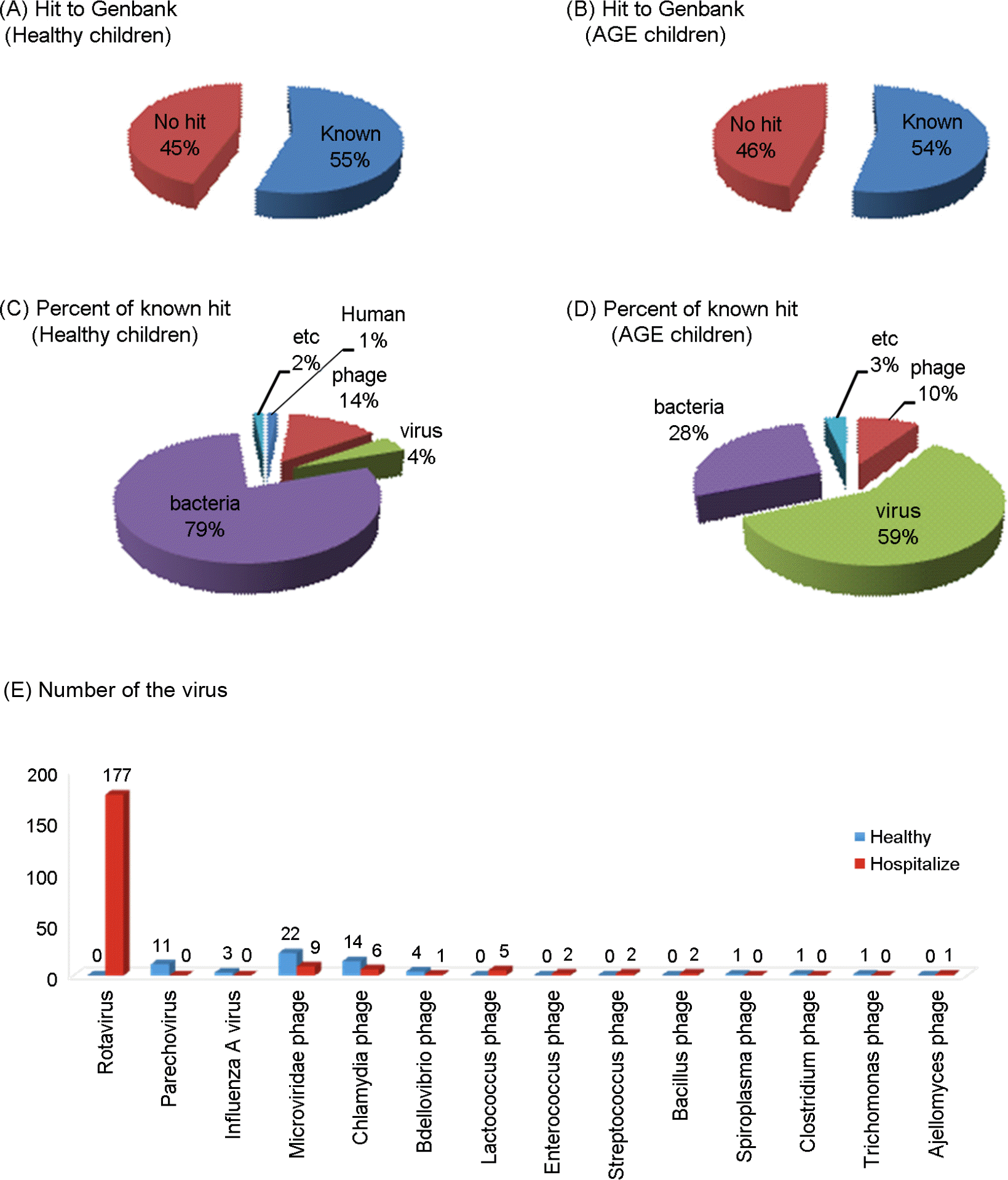 | Figure 1.Genomic overview of the human fecal RNA viruses based on BLAST sequence similarities. Numbers of sequences with significant matches (e values of 0.001) in GenBank (A and C). Distribution of significant matches among known sequences of biological entities (B and D). Distribution of the number of viruses detected in both healthy and hospitalized children (E). |
Table 1.
Genes observed among the virus and phage sequences based on GenBank annotation




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download