Abstract
Helicobacter pylori, a causative agent of gastroduodenal diseases, is a Gram-negative microaerophilic bacterium. Although H. pylori locates in the microaerophilic mucous layer, the bacteria would come into contact harmful reactive oxygen species generated by host immune system. It has been reported that H. pylori harbors various defense mechanisms which can protect bacterial cells from oxygen exposure. The change of the gene expression profile of sodB-negative isogenic mutant of H. pylori 26695 was analyzed by high resolution 2-DE followed by matrix assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) and tandem MS and microarray analysis. Eighteen genes and 41 genes were upregulated and downregulated respectively, either transcriptionally or translationally. Expression levels of three genes including trxB, yxjE and ribE that were changed both on a mRNA level and on a protein level were confirmed by RT-PCR analysis. However, change of expression levels of other major antioxidants such as KatA, AhpC and NapA were not detected, which means Sod is regulated by different way from that of KatA and AhpC. Mutant study of other antioxidant proteins may give us better understanding for the regulation of stress response in H. pylori.
REFERENCES
1). Marshell BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984; 16:1311–5.
2). International Agency for Research on Cancer. Schistosomes, liver flukes and Helicobacter pylori. IARC monogr eval cacinog risks hum. 1994; 61:1–241.
3). Seo JH, Koo SI, Youn HS, Jun JS, Lim JY, Park CH, et al. Comparison of the antibiotic resistance of Helicobacter pylori isolated in Jinju over a 15-year period. J Bacteriol Virol. 2012; 42:305–12.
4). Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997; 388:539–47.
5). Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, et al. Genomic sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999; 397:176–80.
6). Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999; 21:33–7.

7). Bukau B. Regulation of the Escherichia coli heat-shock response. Mol Microbiol. 1993; 9:671–80.
8). Park JW, Song JY, Hwang HR, Park HJ, Youn HS, Seo JH, et al. Proteomic analysis of thiol-active proteins of Helicobacter pylori 26695. J Bacteriol Virol. 2012; 42:211–23.
9). Homuth G, Domm S, Kleiner D, Schumann W. Transcriptional analysis of major heat shock genes of Helicobacter pylori. J Bacteriol. 2000; 182:4257–63.
10). Huesca M, Goodwin A, Bhagwansingh A, Hoffman P, Lingwood CA. Characterization of an acidic-pH-inducible stress protein (hsp70), a putative sulfatide binding adhesin, from Helicobacter pylori. Infect Immun. 1998; 66:4061–7.
11). Matsukawa Y, Asai Y, Kitamura N, Sawada S, Kurosaka H. Exacerbation of rheumatoid arthritis following Helicobacter pylori eradication: disruption of established oral tolerance against heat shock protein? Med Hypotheses. 2005; 64:41–3.
12). Chuang MH, Wu MS, Lin JT, Chiou SH. Proteomic analysis of proteins expressed by Helicobacter pylori under oxidative stress. Proteomics. 2005; 5:3895–901.
13). Seyler RW Jr, Olson JW, Maier RJ. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect Immun. 2001; 69:4034–40.
14). Joo JS, Park KC, Song JY, Kim DH, Lee KJ, Kwon YC, et al. A thin-layer liquid culture technique for the growth of Helicobacter pylori. Helicobacter. 2010; 15:295–302.
15). Park JW, Lee KJ, Kim KM, Joo JS, Kwon YC, Youn HS, et al. Analysis of low molecular weight proteome from H. pylori cell extract using the high performance liquid chromatography. J Bacteriol Virol. 2010; 40:67–75.
16). Alting-Mees MA, Short JM. pBluescript II: gene mapping vectors. Nucleic Acids Res. 1989; 17:9494.

17). Song JY, Park SG, Kang HL, Lee WK, Cho MJ, Park JU, et al. pHP489, a Helicobacter pylori small cryptic plasmid, harbors a novel gene coding for a replication initiation protein. Plasmid. 2003; 50:236–41.
18). Wang Y, Roos KP, Taylor DE. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J Gen Microbiol. 1993; 139:2485–93.
19). O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975; 250:4007–21.
20). Heukeshoven J, Dernick R. Improved silver staining procedure for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis. 1988; 9:28–32.

21). Kim KM, Lee SG, Joo JS, Kwon YC, Bae DW, Song JY, et al. Proteomic analysis of Helicobacter pylori J99 outer membrane protein by tandem mass spectrometry. J Bacteriol Virol. 2008; 38:53–60.
22). Chuang SE, Daniels DL, Blattner FR. Global regulation of gene expression in Escherichia coli. J Bacteriol. 1993; 175:2026–36.
23). Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997; 278:631–7.

24). Wang G, Alamuri P, Maier RJ. The diverse antioxidant systems of Helicobacter pylori. Mol Microbiol. 2006; 61:847–60.
25). Olczak AA, Seyler RW Jr, Olson JW, Maier RJ. Association of Helicobacter pylori antioxidant activities with host colonization proficiency. Infect Immun. 2003; 71:580–3.
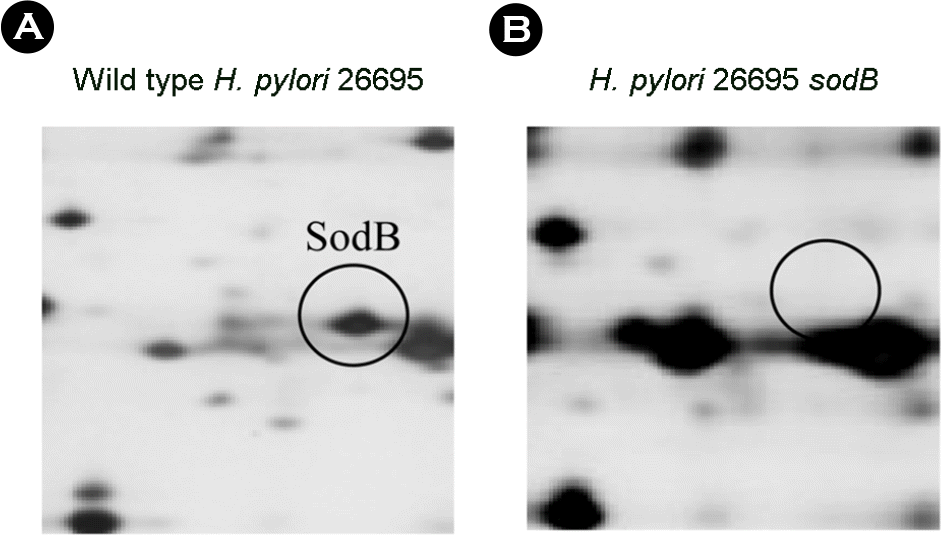
Figure 4.
Comparative proteome analysis of wild type and sodB isogenic mutant of H. pylori 26695. (A) wild type (B) sodB mutant. Numbers on the gel are spot numbers.
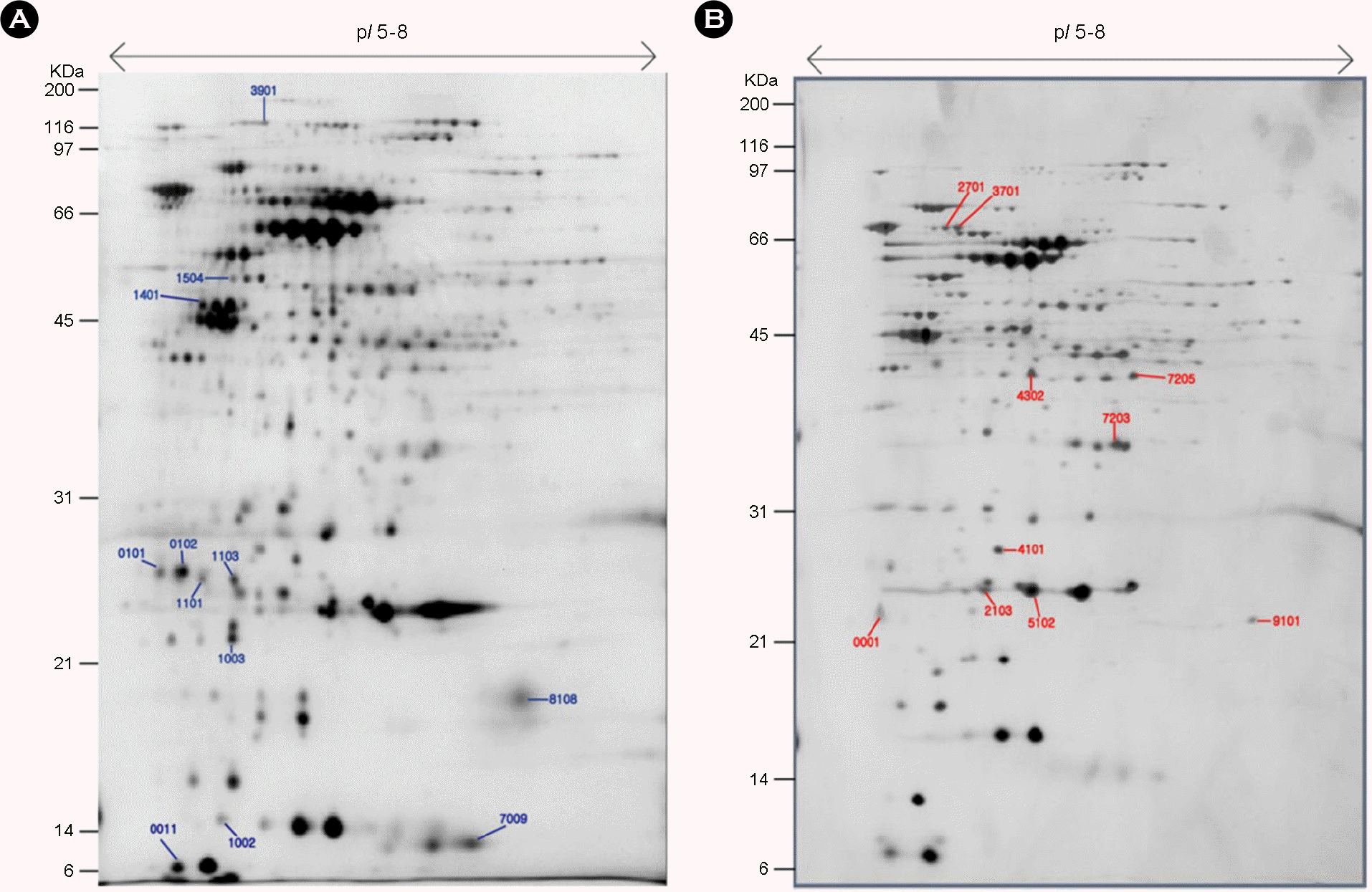
Figure 5.
Comparison of expression of genes in wild type and sodB mutant. (A) RT-PCR analysis of trxB, yxjE, ribE (B) (C) (D) expression of three genes. Expression levels were measured by either 2-DE analysis or microarray. RT-PCR was used to confirm the results.
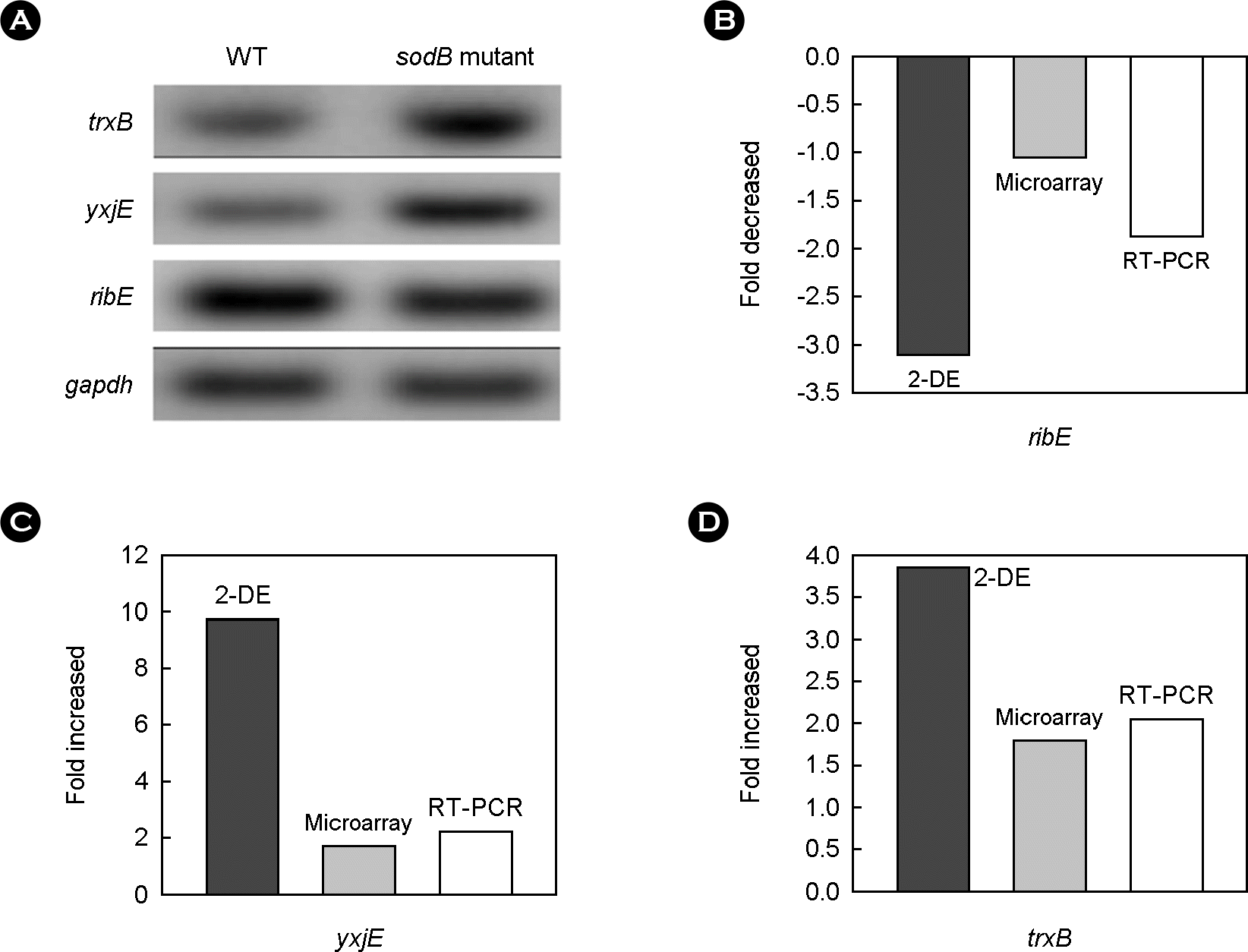
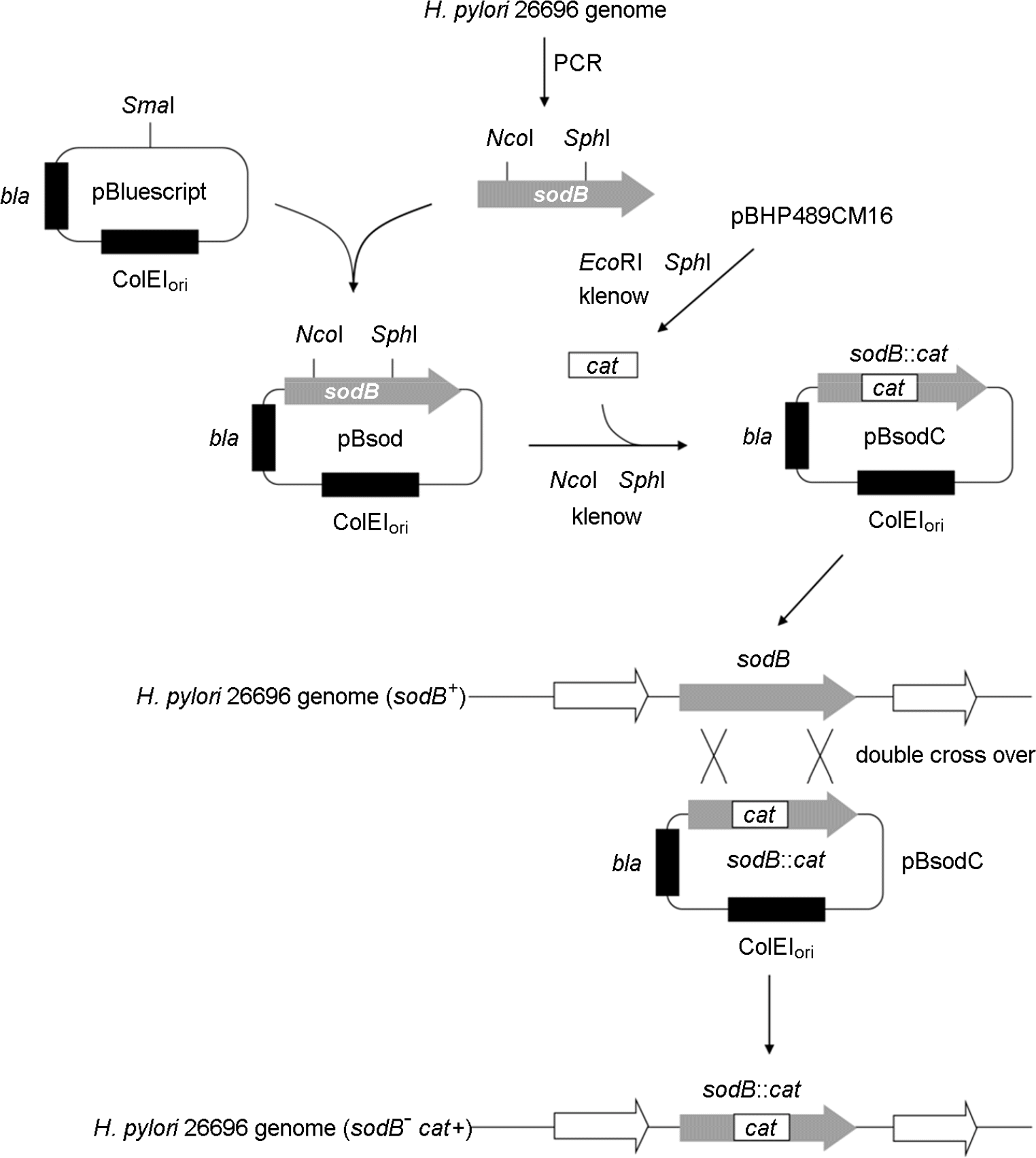
Table 1.
Strains and plasmids used in this study
Table 2.
Lists of up or down-regulated proteins of H. pylori 26695 sodB defective mutant identified by MALDI-TOF
Table 3.
Lists of up or down-regulated proteins of H. pylori 26695 sodB-defective mutant identified by tandem MS
Table 4.
List of genes of which expression is increased or decreased over 2-fold in sodB isogenic mutant compared with H. pylori 26695
Table 5.
Clusters of orthologous groups of genes (COGs) of 24 genes 2-fold changes in expression levels of sodB mutants compared with wild type H. pylori 26695.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download