Abstract
Acinetobacter baumannii is a gram-negative organism reported worldwide as a cause of health-care associated infections. Due to its increasing drug resistance, several studies on coproduction of armA and carbapenemase in South Korea and other parts of the world were reported, which can pose significant therapeutic threat. The aim of this study was to investigate genetic characteristics of multidrug-resistant A. baumannii coproducing armA and carbapenemase and its epidemiological relatedness. Forty-five multidrug resistant (MDR) A. baumannii clinical isolates were collected. Antimicrobial susceptibility was determined by agar dilution, Etest and VITEK 2 system. The presence of 16S rRNA methylase and carbapenemase were analyzed by polymerase chain reaction (PCR) and sequencing. Repetitive element palindromic (REP)-PCR was also performed for epidemiologic investigation. All of A. baumannii isolates harbored blaOXA-51-like gene and 10 isolates showed an upstream ISAba1. 36 isolates (80%) showed amplification of OXA-23, all of which except one had an upstream ISAba1. 16S rRNA methylase armA was found in 44 isolates with high level resistance to aminoglycosides. The rate of coproduction was found in 36 isolates (80%). All isolates showed dominant two patterns in REP-PCR profile. The prevalence of MDR A. baumannii coproducing OXA-23 and armA was high, which the rate of blaOXA-23 coproduction was also high.
Go to : 
REFERENCES
1). Towner KJ. Acinetobacter: an old friend, but a new enemy. J Hops Infect. 2009; 73:355–63.
2). Lee JY, Ko KS. Antimicrobial resistance and clones of Acinetobacter species and Pseudomonas aeruginosa. J Bacteriol Virol. 2012; 42:1–8.
3). Playford EG, Craig JC, Iredell JR. Carbapenem-resistant Acinetobacter baumannii in intensive care unit patients: risk factors for acquisition, infection and their consequences. J Hosp Infect. 2007; 65:204–11.
4). García-Garmendia JL, Ortiz-Leyba C, Garnacho-Montero J, Jiménez-Jiménez FJ, Pérez-Paredes C, Barrero-Almodóvar AE, et al. Risk factors for Acinetobacter baumannii nosocomial bacteremia in critically ill patients: a cohort study. Clin Infect Dis. 2001; 33:939–46.
5). Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007; 51:3471–84.
6). Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006; 12:826–36.
7). Marques MB, Brookings ES, Moser SA, Sonke PB, Waites KB. Comparative in vitro antimicrobial susceptibilities of nosocomial isolates of Acinetobacter baumannii and synergistic activities of nine antimicrobial combinations. Antimicrob Agents Chemother. 1997; 41:881–5.
8). Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007; 5:939–51.
9). Doi Y, Arakawa Y. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis. 2007; 45:88–94.

10). Doi Y, Adams JM, Yamane K, Paterson DL. Identification of 16S rRNA methylase-producing Acinetobacter baumannii clinical strains in North America. Antimicrob Agents Chemother. 2007; 51:4209–10.
11). Zhou Y, Yu H, Guo Q, Xu X, Ye X, Wu S, et al. Distribution of 16S rRNA methylases among different species of Gram-negative bacilli with high-level resistance to aminoglycosides. Eur J Clin Microbiol Infect Dis. 2010; 29:1349–53.

12). Doi Y, Yokoyama K, Yamane K, Wachino J, Shibata N, Yagi T, et al. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob Agents Chemother. 2004; 48:491–6.
13). Doi Y, de Oliveira Garcia D, Adams J, Paterson DL. Coproduction of novel 16S rRNA methylase RmtD and metallo-β-lactamase SPM-1 in a panresistant Pseudomonas aeruginosa isolate from Brazil. Antimicrob Agents Chemother. 2007; 51:852–6.
14). Lee H, Koh EM, Kim CK, Yum JH, Lee K, Chong Y. Molecular and phenotypic characteristics of 16S rRNA methylase-producing gram-negative bacilli. Korean J Clin Microbiol. 2010; 13:19–26.

15). Yu YS, Zhou H, Yang Q, Chen YG, Li LJ. Widespread occurrence of aminoglycoside resistance due to armA methylase in imipenem-resistant Acinetobacter baumannii isolates in China. J Antimicrob Chemother. 2007; 60:454–5.
16). Kim JW, Heo ST, Jin JS, Choi CH, Lee YC, Jeong YG, et al. Characterization of Acinetobacter baumannii carrying bla(OXA-23), bla(PER-1), and armA in a Korean hospital. Clin Microbiol Infect. 2008; 14:716–8.
17). Jeong HW, Son BR, Shin DI, Ryu D, Hong SB, Han K, et al. Characterization of Acinetobacter baumannii co-producing carbapenemases OXA-23 and OXA-66, and armA 16S ribosomal RNA methylase at a university hospital in South Korea. Korean J Clin Microbiol. 2011; 14:67–73.
18). Brignate G, Migliavacca R, Bramati S, Motta E, Nucleo E, Manenti M, et al. Emergence and spread of a multidrug-resistant Acinetobacter baumannii clone producing both the carbapenemase OXA-23 and the 16S rRNA methylase armA. J Med Microbiol. 2012; 61:653–61.
19). Karah N, Haldorsen B, Hermansen NO, Tveten Y, Ragnhildstveit E, Skutlaberg DH, et al. Emergence of OXA-carbapenemase and 16S rRNA methylase-producing international clones of Acinetobacter baumannii in Norway. J Med Microbiol. 2011; 60:515–21.
20). Ko KS, Suh JY, Kwon KT, Jung SI, Park KH, Kang CI, et al. High rates of resistance to colistin and polymyxin B in subgroups of Acinetobacter baumannii isolates from Korea. J Antimicrob Chemother. 2007; 60:1163–7.
21). National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically. 6th ed.Approved standard NCCLS document M7-A6. Wayne, PA: CLSI;2003.
22). Lee K, Yong D, Jeong SH, Chong Y. Multidrug-resistant Acinetobacter spp.: increasingly problematic nosocomial pathogens. Yonsei Med J. 2011; 52:879–91.
23). Mak JK, Kim MJ, Pham J, Tapsall J, White PA. Antibiotic resistance determinants in nosocomial strains of multidrug-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2009; 63:47–54.
24). Sung JY, Kwon KC, Park JW, Kim YS, Kim JM, Shin KS, et al. Dissemination of IMP-1 and OXA type beta-lactamase in carbapenem resistant Acinetobacter baumannii. Korean J Lab Med. 2008; 28:16–23.
25). Bou G, Cerveró G, Dominguez MA, Quereda C, Martínez-Beltrán J. PCR-based DNA fingerprinting (REP-PCR, AP-PCR) and pulsed-field gel electrophoresis characterization of a nosocomial outbreak caused by imipenem- and meropenem-resistant Acinetobacter baumannii. Clin Microbiol Infect. 2000; 6:635–43.
26). Halstead DC, Abid J, Dowzicky MJ. Antimicrobial susceptibility among Acinetobacter calcoaceticus-baumannii complex and Enterobactericeae collected as part of the Tigecycline Evaluation and Surveillance Trial. J Infect. 2007; 55:49–57.
27). Cho YJ, Moon DC, Jin JS, Choi CH, Lee YC, Lee JC. Genetic basis of resistance to aminoglycosides in Acinetobacter spp. and spread of armA in Acinetobacter baumannii sequence group 1 in Korean Hospitals. Diagn Microbiol Infect Dis. 2009; 64:185–90.
28). Lee H, Yong D, Yum JH, Roh KH, Lee K, Yamane K, et al. Dissemination of 16S rRNA methylase-mediated highly amikacin-resistant isolates of Klebsiella penumoniae and Acinetobacter baumannii in Korea. Diagn Microbiol Infect Dis. 2006; 56:305–12.
29). Sung JY, Kwon KC, Cho HH, Koo SH. Antimicrobial resistance determinants in imipenem-nonsusceptible Acinetobacter calcoaceticus-baumannii complex isolated in Daejeon, Korea. Korean J Lab Med. 2011; 31:265–70.
30). Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, et al. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett. 2006; 258:72–7.
31). Akers KS, Chaney C, Barsoumian A, Beckius M, Zera W, Yu X, et al. Aminoglycoside Resistance and Susceptibility Testing Errors in Acinetobacter baumanniicalcoaceticus Complex. J Clin Microbiol. 2010; 48:1132–8.
32). Jung S, Yu JK, Shin SH, Park KG, Jekarl DW, Han K, et al. False susceptibility to amikacin by VITEK 2 in Acinetobacter baumannii harboring armA. Ann Clin Lab Sci. 2010; 40:167–71.
Go to : 
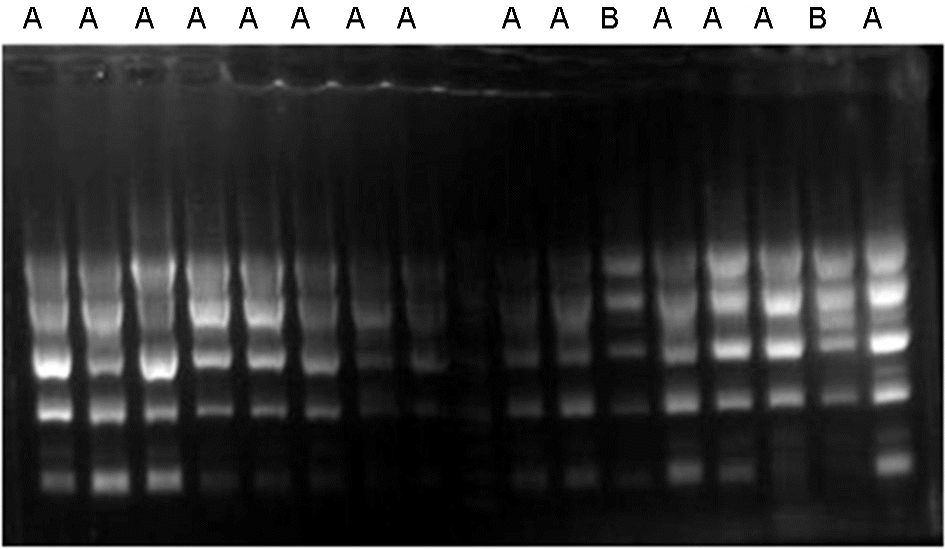 | Figure 1.Repetitive extragenic palindromic (REP)-PCR of genomic DNA from MDR A. baumannii clinical isolates. Most isolates showed A pattern, while some isolate 208 and 218 showed B type. |
Table 1.
Primers used in this study
Table 2.
Minimal inhibitory concentrations of different antimicrobial agents
MICs were determined by Etest (bioMérieux, Marcy l'Etoile, France). But MICs of antimicrobials except Amikacin, gentamicin, tobramycin, imipenem and meropenem were determined by VITEK 2 system (bioMérieux, Marcy l'Etoile, France) in isolates in red color. Abbreviations: MIC, minimal inhibitory concentration; CIP, ciprofloxacin; MN, minocycline; CS, colistin; AN, amikacin; GEN, gentamicin; TOB, tobramycin; IMP, imipenem, MEM, meropenem, FEP, cefepime, P/T, piperacillin/tazobactam; AZ, azteronam.
Table 3.
Genetic characteristics of MDR A. bauamannii




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download