Abstract
The multiple post-translational modifications of proteins display specific gain- or loss-of-function under normal and abnormal conditions. These modifications are precisely regulated by post-translational modification enzymes. The altered molecular status perturbs the pattern of gene expression and decides on a direction to signal transduction cascades as well as intrinsic properties of the proteins. Ultimately, it strictly maintains intracellular environment or results in disease manifestations. Recently, it has become that enzyme-dependent modification of arginine residue to citrulline exerts an important role in the induction of autoimmunity including rheumatoid arthritis, multiple sclerosis, and cancer. The modification of arginine residue to citrulline on proteins is called ‘citrullination’ or ‘deimination’ and is regulated by the calcium-dependent enzyme peptidylarginine deiminase (PAD). Now many effective PAD inhibitors (for example, Cl-amidine) have developed that ameliorates disease phenotypes. In this review, we discuss crucial roles of PAD enzyme and citrullination, the effectiveness of PAD inhibitors, and the implication in pathology.
Go to : 
REFERENCES
1). Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays. 2003; 25:1106–18.

2). Moscarello MA, Mastronardi FG, Wood DD. The Role of Citrullinated Proteins Suggests a Novel Mechanism in the Pathogenesis of Multiple Sclerosis. Neurochem Res. 2007; 32:251–6.

3). Foulquier C, Sebbag M, Clavel C, Chapuy-Regaud S, Al Badine R, Méchin MC, et al. Peptidyl arginine deiminase type 2 (PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in rheumatoid arthritis synovium in close association with tissue inflammation. Arthritis Rheum. 2007; 56:3541–53.

4). Chang X, Zhao Y, Sun S, Zhang Y, Zhu Y. The expression of PADI4 in synovium of rheumatoid arthritis. Rheumatol Int. 2009; 29:1411–6.

5). Chang X, Han J, Pang L, Zhao Y, Yang Y, Shen Z. Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer. 2009; 9:40.

7). Rogers GE, Taylor LD. The enzymic derivation of citrulline residues from arginine residues in situ during the biosynthesis of hair proteins that are cross-linked by isopeptide bonds. Adv Exp Med Biol. 1977; 86A:283–94.

8). Takahara H, Okamoto H, Sugawara K. Calcium-dependent properties of peptidylarginine deiminase from rabbit skeletal muscle. Agric Biol Chem. 1986; 50:2899–904.

9). Senshu T, Kan S, Ogawa H, Manabe M, Asaga H. Preferential deimination of keratin K1 and filaggrin during the terminal differentiation of human epidermis. Biochem Biophys Res Commun. 1996; 225:712–9.

10). Nachat R, Méchin MC, Takahara H, Chavanas S, Charveron M, Serre G, et al. Peptidylarginine deiminase isoforms 1–3 are expressed in the epidermis and involved in the deimination of K1 and filaggrin. J Invest Dermatol. 2005; 124:384–93.

11). Kizawa K, Takahara H, Troxler H, Kleinert P, Mochida U, Heizmann CW. Specific citrullination causes assembly of a globular S100A3 homotetramer: a putative Ca2+ modulator matures human hair cuticle. J Biol Chem. 2008; 283:5004–13.
12). Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Structural basis for Ca(2+)-induced activation of human PAD4. Nat Struct Mol Biol. 2004; 11:777–83.

13). Yao H, Li P, Venters BJ, Zheng S, Thompson PR, Pugh BF, et al. Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis. J Biol Chem. 2008; 283:20060–8.

14). Li P, Yao H, Zhang Z, Li M, Luo Y, Thompson PR, et al. Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol Cell Biol. 2008; 28:4745–58.

15). Li P, Wang D, Yao H, Doret P, Hao G, Shen Q, et al. Coordination of PAD4 and HDAC2 in the regulation of p53-target gene expression. Oncogene. 2010; 29:3153–62.

16). Guo Q, Fast W. Citrullination of inhibitor of growth 4 (ING4) by peptidylarginine deminase 4 (PAD4) disrupts the interaction between ING4 and p53. J Biol Chem. 2011; 286:17069–78.

17). Tanikawa C, Espinosa M, Suzuki A, Masuda K, Yamamoto K, Tsuchiya E, et al. Regulation of histone modification and chromatin structure by the p53-PADI4 pathway. Nat Commun. 2012; 3:676.

18). Wang Y, Li M, Stadler S, Correll S, Li P, Wang D, et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. 2009; 184:205–13.

19). Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med. 2010; 207:1853–62.

20). Zhang X, Bolt M, Guertin MJ, Chen W, Zhang S, Cherrington BD, et al. Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor α target gene activation. Proc Natl Acad Sci U S A. 2012; 109:13331–6.

21). Wright PW, Bolling LC, Calvert ME, Sarmento OF, Berkeley EV, Shea MC, et al. ePAD, an oocyte and early embryo-abundant peptidylarginine deiminase-like protein that localizes to egg cytoplasmic sheets. Dev Biol. 2003; 256:73–88.

22). Yurttas P, Vitale AM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA, et al. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008; 135:2627–36.

23). Jang B, Jin JK, Jeon YC, Cho HJ, Ishigami A, Choi KC, et al. Involvement of peptidylarginine deiminase-mediated posttranslational citrullination in pathogenesis of sporadic Creutzfeldt-Jakob disease. Acta Neuropathol. 2010; 119:199–210.

24). Ishigami A, Ohsawa T, Hiratsuka M, Taguchi H, Kobayashi S, Saito Y, et al. Abnormal accumulation of citrullinated proteins catalyzed by peptidylarginine deiminase in hippocampal extracts from patients with Alzheimer's disease. J Neurosci Res. 2005; 80:120–8.

25). Klareskog L, Rönnelid J, Lundberg K, Padyukov L, Alfredsson L. Immunity to citrullinated proteins in rheumatoid arthritis. Annu Rev Immunol. 2008; 26:651–75.

26). Méchin MC, Sebbag M, Arnaud J, Nachat R, Foulquier C, Adoue V, et al. Update on peptidylarginine deiminases and deimination in skin physiology and severe human diseases. Int J Cosmet Sci. 2007; 29:147–68.

27). Chumanevich AA, Causey CP, Knuckley BA, Jones JE, Poudyal D, Chumanevich AP, et al. Suppression of colitis in mice by Cl-amidine: a novel peptidylarginine deiminase inhibitor. Am J Physiol Gastrointest Liver Physiol. 2011; 300:G929–38.

28). Slack JL, Causey CP, Thompson PR. Protein arginine deiminase 4: a target for an epigenetic cancer therapy. Cell Mol Life Sci. 2011; 68:709–20.

29). Harre U, Georgess D, Bang H, Bozec A, Axmann R, Ossipova E, et al. Induction of osteoclastogenesis and bone loss by human autoantibodies against citrullinated vimentin. J Clin Invest. 2012; 122:1791–802.

30). McGraw WT, Potempa J, Farley D, Travis J. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infect Immun. 1999; 67:3248–56.
31). Abdullah SN, Farmer EA, Spargo L, Logan R, Gully N. Porphyromonas gingivalis peptidylarginine deiminase substrate specificity. Anaerobe. 2013. doi: 10.1016/j.anaerobe.2013.07.001.
32). Wegner N, Wait R, Sroka A, Eick S, Nguyen KA, Lundberg K, et al. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and α-enolase: implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 2010; 62:2662–72.
33). Luo Y, Arita K, Bhatia M, Knuckley B, Lee YH, Stallcup MR, et al. Thompson PR. Inhibitors and inactivators of protein arginine deiminase 4: functional and structural characterization. Biochemistry. 2006; 45:11727–36.
34). Moscarello MA, Lei H, Mastronardi FG, Winer S, Tsui H, Li Z, et al. Inhibition of peptidyl-arginine deiminases reverses protein-hypercitrullination and disease in mouse models of multiple sclerosis. Dis Model Mech. 2013; 6:467–78.

35). Wang Y, Li P, Wang S, Hu J, Chen XA, Wu J, et al. Anticancer peptidylarginine deiminase (PAD) inhibitors regulate the autophagy flux and the mammalian target of rapamycin complex 1 activity. J Biol Chem. 2012; 287:25941–53.

36). Willis VC, Gizinski AM, Banda NK, Causey CP, Knuckley B, Cordova KN, et al. N-α-benzoyl-N5-(2-chloro-1-iminoethyl)-L-ornithine amide, a protein arginine deiminase inhibitor, reduces the severity of murine collagen-induced arthritis. J Immunol. 2011; 186:4396–404.

37). Knight JS, Zhao W, Luo W, Subramanian V, O'Dell AA, Yalavarthi S, et al. Peptidylarginine deiminase inhibition is immunomodulatory and vasculoprotective in murine lupus. J Clin Invest. 2013; 123:2981–93.

38). Meuth SG, Göbel K, Wiendl H. Immune therapy of multiple sclerosis–future strategies. Immune therapy of multiple sclerosis–future strategies. Curr Pharm Des. 2012; 18:4489–97.
39). Arandjelovic S, McKenney KR, Leming SS, Mowen KA. ATP induces protein arginine deiminase 2-dependent citrullination in mast cells through the P2X7 purinergic receptor. J Immunol. 2012; 189:4112–22.

40). Lee HJ, Joo M, Abdolrasulnia R, Young DG, Choi I, Ware LB, et al. Peptidylarginine deiminase 2 suppresses inhibitory {kappa}B kinase activity in lipopolysaccharide-stimulated RAW 264.7 macrophages. J Biol Chem. 2010; 285:39655–62.
42). Sokolove J, Zhao X, Chandra PE, Robinson WH. Immune complexes containing citrullinated fibrinogen costimulate macrophages via Toll-like receptor 4 and Fcγ receptor. Arthritis Rheum. 2011; 63:53–62.

44). Jang B, Jeon YC, Choi JK, Park M, Kim JI, Ishigami A, et al. Peptidylarginine deiminase modulates the physiological roles of enolase via citrullination: links between altered multifunction of enolase and neurodegenerative diseases. Biochem J. 2012; 445:183–92.

Go to : 
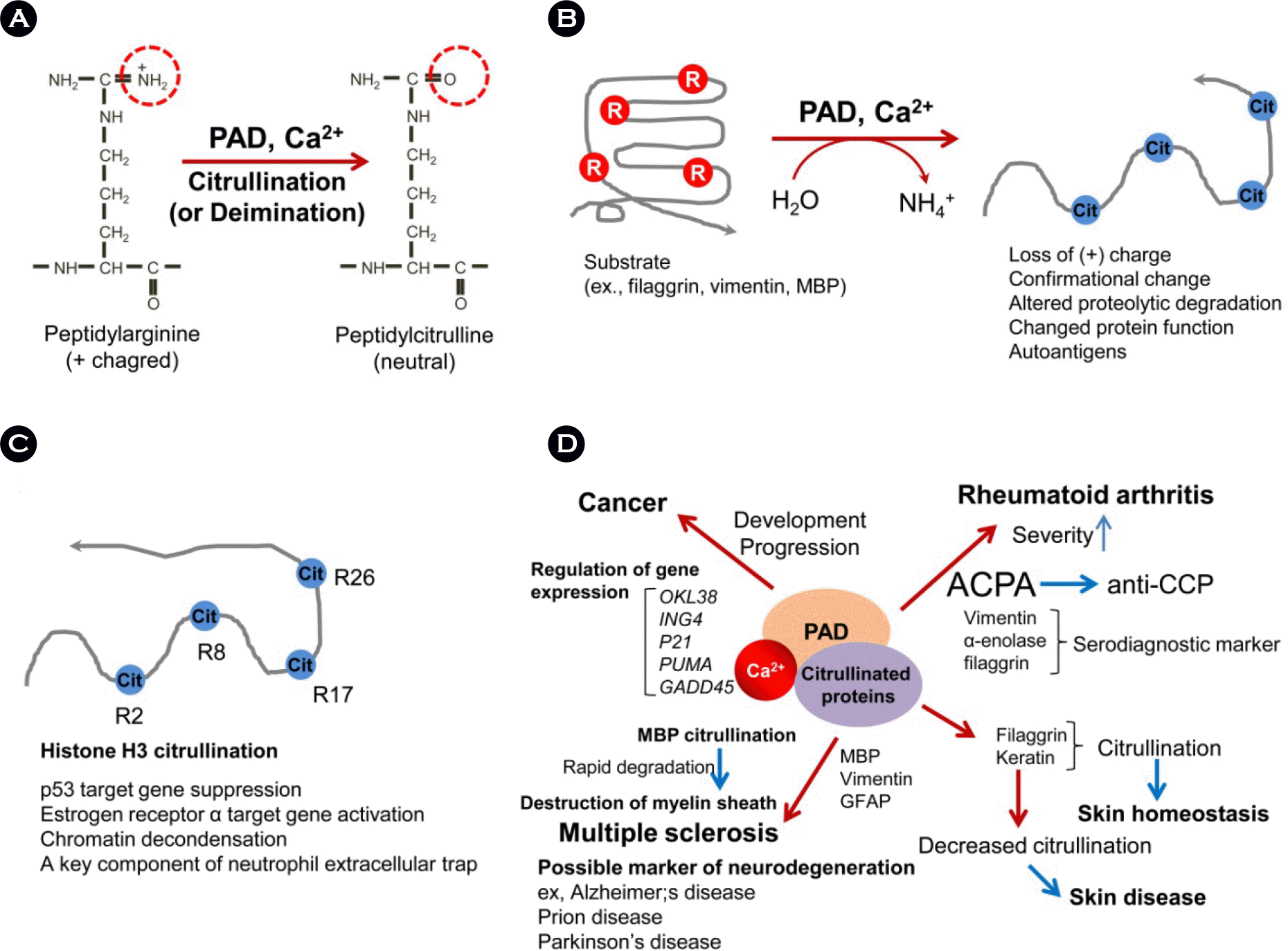 | Figure 1.Peptidylarginine deiminase (PAD) and citrullination. (A) The conversion of peptidylarginine to peptidylcitrulline is a posttranslational process driven by peptidylarginine deiminase (PAD) in a calcium (Ca2+) dependent manner. Ca2+ is an essential cofactor for the activation of PAD, effective Ca2+ concentration is approximately 100-fold higher than physiological cytosolic Ca2+ concentration. (B) Citrullination loss of target positive charge, alters protein structure and susceptibility of protease, changes natural functions, and become autoantigen. (C) Histone citrullination is involved in chromatin decondensation and formation of neutrophil extracellular traps. Citrullinated histones regulate gene transcription including p53- and estrogen receptor α-targeted. (D) PAD and citrullinated proteins are implicated in human diseases including cancer, rheumatoid arthritis, skin physiology, and neurodegeneration. |
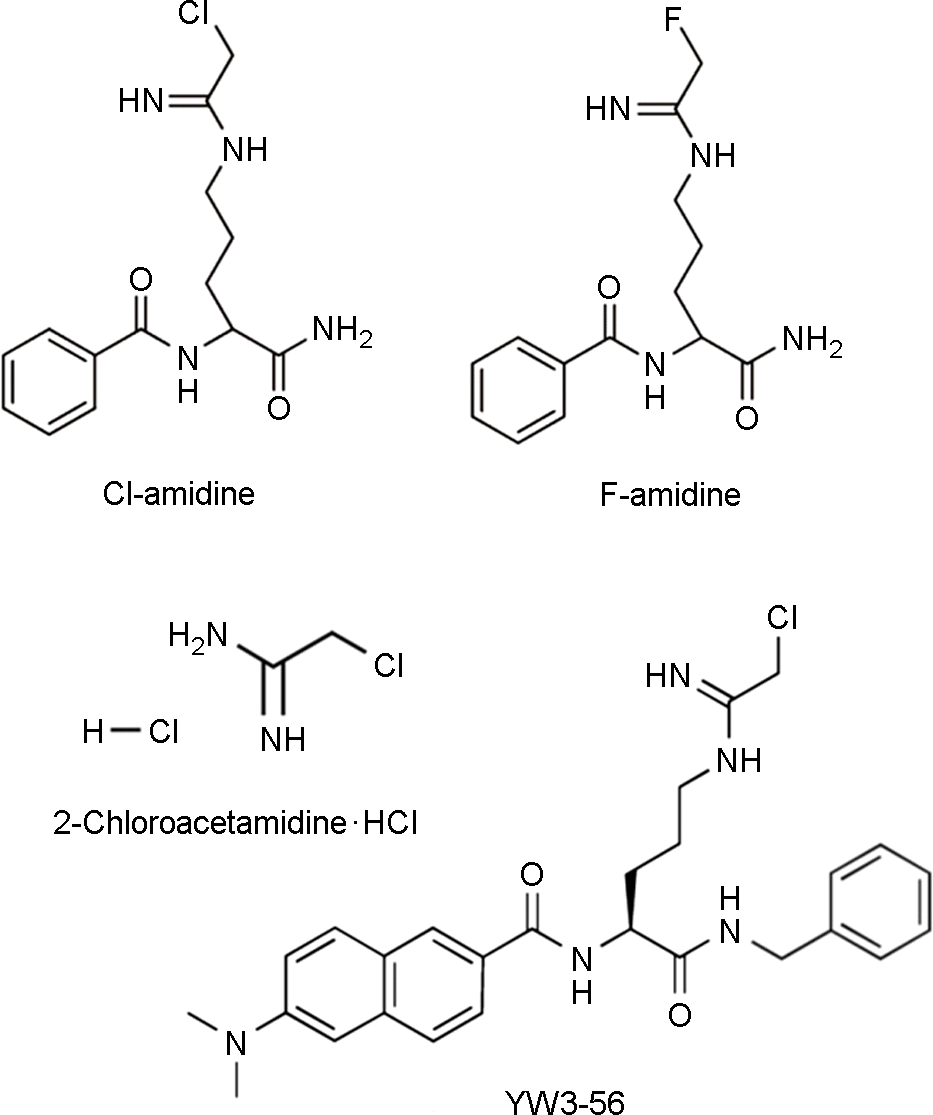 | Figure 2.Structures of Cl- and F-amidine, 2-Chloroacetamidine, and YW3-56. Each PAD inhibitors irreversibly inactivates PAD enzymes via the specific modification of Cys645, an active site residue that is critical for catalysis. |
Table 1.
Peptidylarginine deiminase: protein expression and related diseases.
Table 2.
Potent PAD inhibitors and in vivo test




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download