Abstract
Turnip yellow mosaic virus (TYMV) is a non-enveloped icosahedral virus that has a single 6.3 kb positive-strand RNA as a genome. Previously, it was observed that the recombinant construct TY-eGFP2, where an eGFP gene was inserted at the position downstream of the coat protein (CP) ORF of TYMV genome, barely replicated. The inhibition of replication was relieved by insertion of an additional copy of the 3' quarter of the CP ORF after the foreign sequence. In this study, we have examined if the 3' quarter of the CP ORF contains any replication elements. M-fold analysis predicted three stem-loop structures in this region. Analysis of the TY-eGFP2 constructs containing one or two of these stem-loop structures indicates that the secondary structure predicted in the region between nt-6139 and nt-6181, termed SL2, is essential for TYMV replication. The critical role of SL2 was confirmed by the observation that deletion of the 3' quarter of the CP ORF from the wild-type TYMV genome nearly abolished replication and that insertion of SL2 into the deletion mutant restored the replication. Mutations disrupting the stem of SL2 greatly reduced viral RNA replication, indicating that the secondary structure is essential for the enhancing activity.
Turnip yellow mosaic virus (TYMV) is a positive-strand RNA virus and the type member of the Tymovirus genus. TYMV is a non-enveloped icosahedral virus that contains a monopartite 6.3 kb genomic RNA (gRNA) that encodes p206 (replicase), p69 (movement protein), and coat protein (1). The coat protein is expressed from a subgenomic RNA (sgRNA) that is produced during replication of TYMV. The gRNA and sgRNA have a tRNA-like structure (TLS) at their 3'-ends, which serves as a translation enhancer (2, 3). Two proteins, whose open reading frames (ORFs) extensively overlap, are translated from the genomic RNA. p206, which contains domains for RNA capping, NTPase/helicase and polymerase activities, is the only viral protein necessary for replication in a single cell. It is produced as a polyprotein and self-cleaved to yield 141- and 66-kDa proteins, the latter of which is a RNA-dependent RNA polymerase (RdRP) (1). p69 is required for virus movement within the plant and is also a suppressor of RNA silencing (4).
The information on how TYMV RNA is replicating is limited. Studies of template requirements in vitro using purified TYMV RdRP showed that the enzyme preparation selectively transcribed TYMV RNA to make minus strand RNA. The TYMV RdRP is also used as a template a rather wide range of shorter RNAs that possess -CCA 3'-termini (5). Template copying was predominantly found to be controlled by the -CCA "initiation box" at the 3'-end of the RNA, which needs to be non base-paired and readily accessible (6~8). Although it has been postulated that cis-elements capable of enhancing the specificity and activity of the 3' initiation box exist upstream of the TLS, such replication elements have not been identified by in vitro studies.
In vivo studies have shown existence of replication elements required for TYMV replication. 5'-UTR of TYMV gRNA has two hairpin structures, which had been implicated both in replication and encapsidation of viral RNA. Two hairpins present in TYMV 5'-UTR contain a symmetrical internal loop consisting of C-C or C-A mismatches (9). Mutations with altered C-C and C-A pairs resulted in delay in symptom development and poor encapsidation. The revertants with wild-type phenotype from several passages of infection of the mutants were found to have restored C-C or C-A mismatches, indicating the importance of the mismatches (10). Recently, analysis of viral RNA after agroinfiltration of various 5'-UTR mutants confirmed the involvement of the 5'-UTR in TYMV replication, although the result disproved the role of the two hairpins in TYMV RNA encapsidation. Deletion of one or both hairpins and deletion or mutation of the 17-nucleotide region upstream of the hairpins decreased viral amplification by 10- to 1000-fold (11, 12).
In this study, we report presence of another key sequence element for TYMV replication. The element, presumed to take a stem-loop structure, plays a critical role in TYMV replication, since deletion of this element can nearly abolish viral replication.
TYW and TY-eGFP2 constructs have been described previously (13). The constructs TY-eGFP2+CP6059, TY-eGFP2+CP6139, and TY-eGFP2+CP6186 were made by replacing the sequence between the EcoRI site and XbaI site of the TY-eGFP2 with a PCR-amplified DNA. To make the TY-eGFP2+CP6059 construct, the following primers were used to amplify the sequence from nt-6059 to the 3'-end of the TYW: upstream primer TY(+)6059 (5'-CGAGAATTCAGGCCTCCCGGGTCAAAGATTCGATTCAGT-3'; EcoRI and StuI sites are underlined and TYMV sequence is in italic) and downstream primer HDV-R29 (5'-GGAGAATTCTCTAGATGGCTCTCCCTTAG-3'; EcoRI and XbaI sites are underlined and hepatitis delta virus (HDV) sequence is in italic). The upstream primers used to make the TY-eGFP2+CP6139 and TY-eGFP2+CP6186 were as follows: TY(+)6139 (5'-CGAGAATTCAGGCCTGCATCGACCTGCATAATAACTGTAT-3') and TY(+)6186 (5'-CGAGAATTCAGGCCTTCCGCTCATCACGGACACTTCCACCT-3'). The construct TYΔCP2 was made by replacing the sequence between SmaI and EcoRI sites of the TY-eGFP2+CP6203 with a linker. The oligonucleotides used for the linker are as follows: 5'-CCGGGTCACTAGTACGCGTG-3' and 5'-AATTCACGCGTACTAGTGAC-3'. For making the TYΔCP2+SL2 construct, a linker containing the wild-type SL2 sequence was inserted between the SmaI and EcoRI sites of the TYΔCP2. Other SL2 variant constructs (mSL2, mSL2a, and mSL2b) were prepared likewise. After these manipulations, the resulting constructs were verified by sequence analyses.
Agroinfiltration of the Agrobacterium tumefaciens harboring various TYMV constructs into Nicotiana benthamiana was carried out as described previously (14). Agroinfiltration of all the constructs was accompanied by agroinfiltration of a binary vector supporting the expression of Tomato bushy stunt virus (TBSV) p19 (gene silencing suppressor). Seven days after agroinfiltration, the infiltrated leaves were collected. For RNA and protein extraction, the leaf samples were frozen in liquid nitrogen immediately after collection and stored at -80℃.
Total RNA was isolated from frozen leaf samples using Easy-Red (iNtRON Biotechnology, Sungnam, Korea). RNA samples were resuspended in 48% formamide solution containing 10 mM EDTA and incubated at 65℃ for 10 min before electrophoresis on 1% agarose gel. RNAs were transferred to Hybond N+ membranes (GE Healthcare, Chalfont St. Giles, England), and the blots were hybridized with a digoxigenin (DIG)-labeled DNA probe hybridizing to the TYMV CP ORF, as described previously (14). The blots were developed by chemiluminescent immunodetection of DIG (Roche Molecular Biochemicals, Basel, Switzerland).
Coat protein (CP) expression was examined by 12.5% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blot analysis. Leaf samples (0.1 g) were ground in 200 µl of 2X sample buffer (125 mM Tris-HCl, pH 6.8, 20% glycerol, 4% SDS, 2% 2-mercaptoethanol, 10 µg/ml bromophenol blue) and boiled for 5 min before analysis. Western blot was performed with anti-TYMV rabbit antiserum in conjunction with horseradish peroxidase-conjugated goat anti-rabbit IgG, followed by chemiluminescent detection using Luminata™ Forte (Millipore, Bedford, MA, USA).
Previously, it was observed that the replication of TYMV was abolished when a foreign sequence was inserted between the CP ORF and the 3'-terminal tRNA-like structure. The inhibition was relieved in the construct TY-eGFP2-CP6059 (Fig. 1A), where 3' quarter of CP ORF was additionally inserted at the position downstream of the foreign sequence (13). This suggests that the 3' quarter of the CP ORF contains an essential replication element. Since the TY-eGFP2 recombinants contain an intact CP ORF sequence upstream of the eGFP gene, this result also indicates that the replication element should be appropriately spaced with respect to the 3'-end for its function.
In many positive RNA viruses, the RNA replication elements consisting of secondary structures such as hairpins are scattered on the viral genome, and they interact with each other to enhance or suppress the replication and translation (15). To see if the 3' quarter of the TYMV CP ORF contains any secondary structures, the region was analyzed by the M-fold secondary structure prediction program (16). The result revealed three stem-loop structures between the nt-6059 and the stop codon of the CP ORF (Fig. 1B).
We then examined the role of the stem-loop (SL) structures by inserting one or two of the SL structures into the TY-eGFP2. Analysis of viral RNA accumulation showed that the predicted stem-loop structure with an asymmetric bulge (SL2), present between nt-6139 and nt-6181, was critical to the replication of the recombinant TYMV (Fig. 2). Addition of SL3 alone did not enhance the variant TYMV replication. Insertion of SL2 (TY-eGFP2+CP6139) restored the replication, although the replication was somewhat weaker than the TY-eGFP2+CP6059. This result indicates that the SL1 structure or the sequence between the SL1 and SL2 may have an additional enhancing effect on TYMV replication.
To see whether the results obtained with recombinants can be extended to wild-type TYMV, we made the construct TYΔCP2, where the 3' quarter of CP ORF was deleted (Fig. 3A). TYΔCP2 lacks the sequence between nt-6068 and nt-6202. When the replication of the TYΔCP2 was examined, it was observed that the deletion mutant barely replicated (Fig. 3B). This result again shows that the region lacked in TYΔCP2 contains an essential sequence element for TYMV replication. In a previous study, it was reported that a deletion in CP ORF resulted in decrease in TYMV replication (17). The mutant lacks the sequence between nt-5707 (PvuII site) and nt-6061 (SmaI site), comprising two-thirds of the CP ORF. The replication of the mutant was reported to decrease to the level 10% that of wild-type TYMV. The authors described that the reduced accumulation of viral RNA might be due to increased degradation of the non-encapsidated replication product. Here, we observed that TYMV replication was nearly abolished when the 3' quarter of CP ORF was deleted.
We then examined whether replication could be recovered by insertion of SL2 to the TYΔCP2. In TYΔCP2+SL2, the sequence representing the SL2 was inserted back into the TYΔCP2 (Fig. 3A). As shown in Fig. 3B, replication of the TYΔCP2+SL2 was recovered to 5 to 10% of wild type. This partial recovery of replication could be due to defective CP, as described above. Although the CP produced from the TYΔCP2+SL2 is detected by anti-TYMV CP antibody, it is likely that the CP is not functional because 32 amino acids are missing in the CP produced from TYΔCP2+SL2. To examine further the role of the SL2, modified SL2 sequences were added into the TYΔCP2 (Fig. 4A). In the mSL2 constructs, mutations were introduced with intent to disrupt the stem-loop structure. Northern analysis of viral RNA showed that the three SL2 variants were not efficient to stimulate replication, as shown in Fig. 4B. Although mSL2a sequence was found to enhance the viral replication to some extent, it was less efficient, compared to the enhancing effect by SL2. Overall, these results suggest that a sequence element in the CP ORF is required for efficient TYMV replication, and that the stem-loop structure of the element is important for its function.
In positive-strand RNA viruses, cis-acting elements are involved not only in RNA/RNA interactions but also in binding of viral and cellular proteins during the processes of translation, RNA replication and encapsidation. Most viral, cis-acting elements are located in the highly structured 5'- and 3'-untranslated regions of the genomes but sometimes they also extend into the adjacent or distant regions (15, 18, 19). In this study, a stem-loop structure (SL2) in the CP ORF has been identified as a sequence element required for efficient TYMV replication. Although 5'-UTR is also needed for TYMV replication (11, 12), SL2 seems to play a more critical role in TYMV replication. This is because deletion of the 5'-UTR did not abolish TYMV replication completely (12), whereas little replication was observed when the region containing SL2 was deleted. It was observed that the distance between the 3'-end of the TYMV gRNA and SL2 was critical for efficient replication. Considering that the location of SL2 is proximal to the 3'-end of positive-strand viral RNA, it is likely that the SL2 and a nearby structure (or sequence) at the 3'-end might constitute a structural feature, which is recognized by TYMV RdRP when synthesizing minus strand RNA. An interaction between the SL2 and the 3'-end could also be important.
The results obtained in this study are also consistent with the model that the SL2, possibly together with a nearby structure including the 3'-TLS, serves as a recruitment signal. Positive-strand RNA viruses characteristically replicate within membranous vesicles formed on organelles such as ER, mitochondria, chloroplast, and peroxisome (20). For this reason, recruitment of viral RNA to replication vesicles is crucial for replication. During TYMV infection, p66 localizes to virus-induced chloroplastic membrane vesicles, which are closely associated with TYMV RNA replication (1). Targeting of the p66 to the chloroplast envelope is dependent on the expression of p141, which is targeted to the chloroplast in the absence of other viral factors and induces the clumping of the chloroplasts (21). In a subsequent study, the domain in the p141 interacting with the p66 was mapped to the protease domain (22).
TYMV gRNA should also be recruited to the replication complex. However, it is not known how TYMV gRNA is transported to the replication vesicle. In Bromo mosaic virus (BMV), BMV 1a protein, which contains a helicase-like domain and RNA capping functions, is targeted to ER membranes after recruiting 2a protein and the three BMV gRNAs (B1, B2, and B3) (23, 24). Thus, there is a possibility that the TYMV p141 recruits the gRNA as well as the p66 before localization to the chloroplast outer membranes. However, exact nature of the interaction in the process of TYMV RNA recruitment has not been studied. Whether the 3'-proximal SL2 indeed functions as a recruitment element remains to be elucidated.
Figures and Tables
Figure 1
TYMV constructs. (A) Wild-type and recombinant TYMV constructs. TYW represents a wild-type genome. TY-eGFP2 construct has the enhanced green fluorescent protein gene between the CP ORF and the 3'-terminal TLS. TY-eGFP2+CP6203, where the TYMV sequence downstream of the foreign gene begins with the nt-6203 of the wild type, was observed to show little viral replication (13). (B) Secondary structures predicted at the 3'-end proximal region of the CP ORF. The three stem-loop structures were designated SL1, SL2, and SL3. The two constructs shown below were designed to have all or part of the stem-loop structures. TY-eGFP2+CP6139 and TY-eGFP2+CP6186 have additional CP ORF sequence starting with the nt-6139 and nt-6186, respectively. All these constructs have a 3'-terminal ribozyme sequence derived from hepatitis delta virus (HDV).
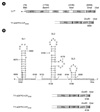
Figure 2
Replication of the TY-eGFP2 constructs having extra CP sequences. (A) Northern and Western analysis of the replication. Seven days after agroinfiltration of N. benthamiana leaf with various TYMV constructs, total RNA was extracted from the leaf. One µg or 0.1 µg (10-1) of total RNA was size-fractionated in the 1% agarose gel and examined by Northern blot analysis, using the DIG-labeled probe representing the CP ORF. The blots were developed by chemiluminescent immunodetection of DIG. The panel below the Northern blot represents a gel stained with ethidium bromide. In Western analysis of coat protein expression, 1 µl of 1:10 diluted (10-1) or undiluted leaf extract was loaded and electrophoresed in 12.5% SDS-polyacrylamide gel. The proteins were transferred to a nitrocellulose membrane. Coat protein was detected using anti-TYMV coat protein rabbit antibody and anti-rabbit HRP conjugate. The membrane was developed by a Luminata™ Forte (Millipore) using luminol as the substrate. (B) Northern analysis using DIG-labeled eGFP sequence as a probe. Note that the smaller sgRNA is evident.
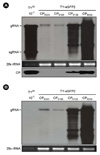
Figure 3
Examination of the role of SL2 in wild-type TYMV. (A) TYMV constructs for dissecting the role of SL2. In TYΔCP2, the nucleotides between #6068 and #6202 were deleted and several cloning sites were added. In TYΔCP2+SL2, the sequence between nt-6137 and nt-6183 was added back to the SmaI and EcoRI sites of the TYΔCP2. (B) Northern and Western analyses of the replication of TYΔCP2 and TY-CP2+SL2 constructs. The analyses were carried out as described in Fig. 2.

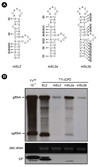
Figure 4
Efficiency of SL2 variants. (A) Constructs containing modified SL2 sequences. The nucleotides in the three TYΔCP2+mSL2 constructs that differ from the wild-type SL2 are highlighted by arrows and grey boxes. Here, only the sequences representing the stem-loop structures are shown. (B) Northern and Western analyses of the replication of various SL2 constructs. The analyses were carried out as described.
References
1. Dreher TW. Turnip yellow mosaic virus: transfer RNA mimicry, chloroplasts and a C-rich genome. Mol Plant Pathol. 2004. 5:367–375.

2. Matsuda D, Dreher TW. The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3'-translational enhancer. Virology. 2004. 321:36–46.

3. Matsuda D, Bauer L, Tinnesand K, Dreher TW. Expression of the two nested overlapping reading frames of Turnip yellow mosaic virus RNA is enhanced by a 5' cap and by 5' and 3' viral sequences. J Virol. 2004. 78:9325–9335.

4. Chen J, Li WX, Xie D, Peng JR, Ding SW. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell. 2004. 16:1302–1313.

5. Singh RN, Dreher TW. Turnip yellow mosaic virus RNA-dependent RNA polymerase: initiation of minus strand synthesis in vitro. Virology. 1997. 233:430–439.

6. Deiman BA, Koenen AK, Verlaan PW, Pleij CW. Minimal template requirements for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J Virol. 1998. 72:3965–3972.

7. Deiman BA, Verlaan PW, Pleij CW. In vitro transcription by the turnip yellow mosaic virus RNA polymerase: a comparison with the alfalfa mosaic virus and brome mosaic virus replicases. J Virol. 2000. 74:264–271.

8. Singh RN, Dreher TW. Specific site selection in RNA resulting from a combination of nonspecific secondary structure and -CCR- boxes: initiation of minus strand synthesis by turnip yellow mosaic virus RNA-dependent RNA polymerase. RNA. 1998. 4:1083–1095.

9. Hellendoorn K, Michiels PJ, Buitenhuis R, Pleij CW. Protonatable hairpins are conserved in the 5'-untranslated region of tymovirus RNAs. Nucleic Acids Res. 1996. 24:4910–4917.

10. Hellendoorn K, Verlaan PW, Pleij CW. A functional role for the conserved protonatable hairpins in the 5' untranslated region of turnip yellow mosaic virus RNA. J Virol. 1997. 71:8774–8779.

11. Shin HI, Cho NJ, Cho TJ. Role of 5'-UTR hairpins of the Turnip yellow mosaic virus RNA in replication and systemic movement. BMB Rep. 2008. 41:778–783.

12. Shin HI, Tzanetakis IE, Dreher TW, Cho TJ. The 5'-UTR of Turnip yellow mosaic virus does not include a critical encapsidation signal. Virology. 2009. 387:427–435.

13. Shin HI, Cho TJ. A sequence in coat protein open reading frame is required for Turnip yellow mosaic virus replication. J Bacteriol Virol. 2011. 41:109–116.

14. Cho TJ, Dreher TW. Encapsidation of genomic but not subgenomic turnip yellow mosaic virus RNA by coat protein provided in trans. Virology. 2006. 356:126–135.

15. Miller WA, White KA. Long-distance RNA-RNA interaction in plant virus gene expression and replication. Annu Rev Phytopathol. 2006. 44:447–467.

16. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003. 31:3406–3415.

17. Weiland JJ, Dreher TW. Cis-preferential replication of the turnip yellow mosaic virus RNA genome. Proc Natl Acad Sci U S A. 1993. 90:6095–6099.

18. Dreher TW. Functions of the 3'-untranslated regions of positive strand RNA viral genomes. Annu Rev Phytopathol. 1999. 37:151–174.

19. Liu Y, Wimmer E, Paul AV. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim Biophys Acta. 2009. 1789:495–517.

20. Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat Rev Microbiol. 2006. 4:371–382.

21. Prod'homme D, Jakubiec A, Tournier V, Drugeon G, Jupin I. Targeting of the turnip yellow mosaic virus 66K replication protein to the chloroplast envelope is mediated by the 140K protein. J Virol. 2003. 77:9124–9135.
22. Jakubiec A, Notaise J, Tournier V, Héricourt F, Block MA, Drugeon G, et al. Assembly of turnip yellow mosaic virus replication complexes: Interaction between the proteinase and polymerase domains of the replication proteins. J Virol. 2004. 78:7945–7957.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download