Abstract
Helicobacter pylori are a capnophilic bacterium, which colonize gastric mucosa and are resistant to acidic and oxidative damage. Thiol-active proteins subserve redox functions in tolerating oxidative stress and environmental toxicants, such as hydrogen peroxide and hypochlorous acid. We analyzed disulfide-containing proteins of H. pylori strain 26695. Active disulfide-containing proteins were separated by thiol-affinity chromatography, displayed with two-dimensional electrophoresis (2-DE), and identified by MALDI-TOF-MS. Thirty-five putative disulfide proteins, including AhpC (HP1563), GroEL (HP0011), and FrdB (HP0191), were identified in this study. In addition, 4 disulfide proteins of HypB, FusA, TufB, and AhpC showed enhanced intensities in the periplasmic space when compared with the pellet, suggesting that these proteins might play roles in the first redox system against environmental oxidative stresses. Disulfide-containing proteins identified in this study will provide the standard landscape for constructing the proteome components responsible for redox regulation of H. pylori.
Helicobacter pylori were first isolated in 1982 by Warren and Marshall (1). H. pylori are gram-negative, spiral-shaped, and capnophilic bacterium, that cause acute and chronic gastritis in humans. More than half of the world's population is infected with H. pylori (2, 3), and although most people infected with H. pylori are asymptomatic or do not develop serious illness (4, 5). However, prolonged exposure of human gastric mucosa to H. pylori has been linked to the development of gastroduodenal ulcers and gastric cancer (6~12). H. pylori has been well known as a microaerobic organism, however, it was recently reported that increased CO2 was required for the growth of H. pylori in vitro, due in part to the presence of carbamoyl-phosphate synthetase (13) and acetyl-CoA carboxylase (14). In addition, the best atmospheric condition for exuberant growth of H. pylori was demonstrated to be a CO2 concentration of 5~10% and an O2 concentration of 18.5~19.4%, meaning that this bacterium is capnophilc, rather than microaerophilic (15). H. pylori is thought to have a variety of redox mechanisms for the defense against oxidants.
Residues of the amino acid cysteine have endowed the proteins with structural, catalytic, and regulatory features due to the nucleophilicity of the thiol functional group (16, 17). By responding to oxidative stress and environmental toxicants, cysteine serves as a key residue in enzyme catalysis (18), protein oxidative folding (19) and trafficking (20, 21), as well as redox signaling and regulation (22~26). The cysteine residue exists in vivo in the fully reduced free thiol form (-SH or -S-) and can oxidized reversibly to the sulfenic (-SOH) form, the thiyl radical (-S•), the disulfide bond (Cys-S-S-Cys), the S-nitrosylated form (-S-NO), or sulfenic esters (-SCO2R) the sulfenyl hydrochloride intermediate (-SCl), and irreversibly to the sulfinic (SO2H) and sulfonic (-SO3H) acid forms (16, 17, 27).
Under the oxidative stress response, reduced free thiol forms of cysteine residues are oxidized to sulfenic acid and are then rapidly condensed to form disulfide bonds through intra- and intermolecular interaction or glutathionylation (17). Over the past years, disulfide containing proteins have been reported from various samples, Dsb family in prokaryotes (28, 29) and PDI family in the endoplasmic reticulum (ER) (30, 31) that catalyze oxidative protein folding in cells. PDI is oxidatively recycled by the ER oxidoreductin proteins Ero1 systems, AERO systems, and Erv2p which couple disulfide bond formation to reduction of molecular oxygen in the presence of FAD cofactor (32). These folding factors are resided in the ER through which a number of cell-surface and secretory proteins are passed on the way to maturity. Disulfide bonds of Yap1 and OxyR were formed in the redox status in vivo and then gave a signal to induce the appropriate level and duration of specific transcription (33, 34). Disulfide bonds might be a post-translational modification system capable of being recycled by redox status and playing a regulatory role in enzymatic activity, signal transduction, transcriptional activity, and protein folding (35).
Global identification of reactive cysteine disulfide containing proteins in an organism was needed to grasp comprehensively the systems responsible for its physiological activities in response to the cytosolic redox potential. Disulfide proteome analysis of various organisms have supported that a wide spectrum of enzymes involved in cellular processes, including metabolic and biosynthetic pathways, energy transduction, transcription factors, and ribosomal protein, could be controlled by a mechanism dependant on the redox state of cells as well as antioxidant systems and regulatory factors (36, 42~51). However, limited bacterial species have been attempted to globally analyze the reactive disulfide proteomes to date (37~42).
Various approaches have been introduced for trapping the cysteine redox state, thiol labeling, and enrichment of reactive cysteine containing proteins (36). All methods have been reviewed to have advantages and drawbacks for purification of reactive disulfide proteins. Here, a total cell lysate of H. pylori 26695 was alkylated and reduced, and applied on the thiol-affinity chromatography to select the reactive disulfide proteins. Bound proteins on the column were eluted by DTT, displayed on 2-DE, and subjected to the peptide (fingerprinting) using MALDI-TOF MS, to identify 35 kinds of reactive disulfide proteins which are involved in various cellular functions. These data supported that disulfide proteome may provide cellular factors which aid intracellular mechanisms of H. pylori in response to redox signaling.
H. pylori strain 26695 was supplied from H. pylori Korean Type Culture Collection (Gyeongsang National University School of Medicine). The frozen H. pylori were thawed and grown on brucella agar plates containing 10% bovine serum, vancomycin (10 µg/ml), nalidixic acid (25 µg/ml), and amphotericin B (1 µg/ml) at 37℃ under 10% CO2 and 100% humidity as described previously (52). The strains were subcultured on a brucella agar plate supplemented with 10% bovine serum and cultured under the same conditions. The strains were daily subcultured on new agar plates until obtaining a homogenous bacterial growth. Bacterial masses were collected for experiments after examining whether most bacteria showed a typically curved shape in the microscopic observation using Gram staining.
Preparation of disulfide containing proteins was carried out as described previously (53). Total proteins from the bacteria were obtained by suspending the cells in 20 ml of 100 Tris/sodium dodecyl sulfate (SDS) buffer (100 mM Tris-HCl, pH 7.5, 1% w/v sodium dodecyl sulfate (SDS), disrupted by the use of an ultrasonicator (Sonics & Materials, Danbury, CN, USA). For protein precipitation, 10% w/v trichloroacetic acid (TCA) was added, and incubation took place at 4℃ for 30 min. Insoluble material containing the protein fraction was collected by centrifugation (8,000 rpm, 15 min), after which it was washed twice with 20 ml cold acetone and dried. The dried pellet was dissolved in 16 ml alkylation cocktail (100 mM Tris-HCl, pH 7.5, 1% w/v SDS, 40 mM iodoacetamide, 40 mM N-ethylmaleimide). Insoluble material was removed after centrifugation (8,000 rpm, 15 min). Alkylation was performed by boiling for 3 min and then incubating for 1 h at 37℃. After alkylation, proteins were precipitated by addition of the same volume of 20% w/v TCA and collected by centrifugation as described above. The TCA pellet was washed twice with cold acetone and was dried. For reduction of active thiol disulfide, the pellet was redissolved in 4 ml Tris/SDS buffer (100 mM Tris-HCl, pH 7.5, 1% w/v SDS) and 40 µl of 400 mM tributylphosphine in dimethylformamide was added, to obtain a final concentration of 4% mM tributylphosphine. After incubation for 20 min at room temperature, proteins were precipitated by adding 4 ml of 20% w/v TCA, and were recovered by centrifugation and washed twice with acetone.
We performed thiol-affinity chromatography as described previously (53). Alkylation and reduction of the sample was applied onto a 2 ml activated thiol-sepharose 4B (Sigma-Aldrich Korea) resin pre-equilibrated with Tris/SDS buffer. The column was washed twice with 6 ml of the same buffer. The bound proteins were eluted with 2 ml Tris/SDS buffer containing 100 mM dithiothreitol (DTT).
The periplasmic fraction of H. pylori cells were prepared by osmotic shock treatment as previously described (54). H. pylori were grown to 1.0~1.3 OD600 with brucella broth supplemented with 10% horse serum in thin layer-liquid culture, and were harvested by centrifugation at 3,000 rpm for 20 min. One OD600-unit of Hp suspension was washed with 1 ml 10 mM Tris-HCl (pH 8) and the bacteria were resuspended in 25% sucrose, 1 mM EDTA, 10 mM Tris-HCl (pH 8). After vigorous shaking for 10 min at room temperature, the bacteria were harvested as above, resuspended quickly in ice cold H2O, and were again shaken for 10 min at 4℃. The supernatant containing periplasmic proteins was collected as the periplasmic fraction and was filtered with 0.2 mm nitrocellulose paper after which the pellet of osmotic shocked bacteria was resuspended in 100 ml PBS and harvested by centrifugation as the precipitant body.
Eluted disulfide protein was cleaned to remove contaminants with TCA precipitation and a 2-D clean up kit (Amersham Biosciences Korea). Two-DE was carried out as described previously (55). The solubilized protein sample (200 µg) was mixed with the rehydration solution containing 8 M urea, 4% CHAPS, 10 mM DTT, and 0.2% carrier ampholytes (pH 5.0~8.0 and 6.0~11.0) to a final volume of 300 µl, and was applied to immobilized pH gradient (IPG) strips (17 cm; Bio-Rad, Hercules, CA, USA) of pH 5.0~8.0 and 6.0~11.0 in a reswelling tray (Bio-Rad). After the IPG strips were rehydrated, IEF was performed using a Protein IEF Cell (Bio-Rad) and three preset programs consisting of the first conditioning step (15 min at 250 V), the linear voltage ramping step (3 h at 10,000 V), and the maximum voltage ramping step of up to 90,000 Vh. The current did not exceed 50 µA per strip. Following IEF, the strips were equilibrated with 0.375 M Tris buffer (pH 8.8) containing 6 M urea, 2% SDS, 20% glycerol, 2% DTT, and 0.01% bromophenol blue. The strips were equilibrated again with the same buffer supplemented with 2.5% iodoacetamide. The second dimension SDS-PAGE was carried out overnight at 20 mA per gel using a 13% separating polyacrylamide gel (18 × 20 cm) without a stacking gel. The resolved protein spots on the gels were visualized by silver staining (55) and scanned using a Fluor-S MultiImager (Bio-Rad). Spot intensities of each sample were documented and analyzed using the PDQUEST 2-D Gel Analysis Software Version 7 (Bio-Rad) installed on a Magic Station M5660 (Samsung, Korea).
The silver-stained spots were excised from the 2-DE gels and were transferred into microcentrifuge tubes. The spots were de-stained with fresh chemical reducers in a 1:1 ratio of 30 mM potassium ferricyanide and 100 mM sodium thiosulfate, as described previously (55, 56), with occasional mixing until the brownish color disappeared. The gel pieces were rinsed three times with distilled water to stop the reaction. Ammonium bicarbonate (500 µl of 200 mM) was added to cover the gels for 20 min, and was then discarded. The gel pieces were dehydrated with 100 µl of acetonitrile and dried in a vacuum centrifuge. An in-gel digestion was carried out by the method described by O'Connell and Stults (57). Gel pieces were rehydrated by covering with digestion buffer containing trypsin (12.5 ng/ml) and incubating on ice for 45 min. The enzyme solution was replaced with 20 µl of the buffer without enzyme and incubated overnight at 37℃. The gel pieces were vortexed vigorously for 30 min, after which 20 µl of the digested solution was transferred into a clean microcentrifuge tube and dried in a vacuum. The resulting pellets were dissolved in 2 µl of 0.1% TFA.
For the matrix solution, α-cyano-4-hydroxycinnamic acid (40 mg/ml) was dissolved in 50% acetonitrile and 0.1% TFA. The matrix and sample solutions (10 µl each) were mixed and loaded into the target wells, rapidly dried, and washed using deionized water. The wells were dried for 10 min at room temperature and subjected to MALDI-TOF-MS analysis using a Voyager Biospectrometry Workstation (PE Biosystems, Foster City, CA, USA) with the following parameters: 20 kV accelerating voltage, 75% grid voltage, 0.02% guide wire voltage, 70 ns delay, and a mass gate from 800 to 2500. The peptide mass fingerprints were analyzed using the program MS-FIT of ProteinProspect developed by the UCSF Mass Spectrometry Faculty (http://prospector.ucsf.edu). The NCBI database of Helicobacter proteins was searched to identify the proteins, using monoisotopic peptide masses and allowing a molecular mass range of 2-DE ± 15%, a peptide mass accuracy of 50 ppm, and one partial cleavage. When matching proteins were not found, the molecular mass window was extended. Pyroglutamic acid modification of N-terminal glutamine, oxidation of methionine, and acrylamide modification of cysteine were taken into consideration.
The whole cells of H. pylori strain 26695 were disrupted by ultrasonic dismembrator. The total proteins of H. pylori strain 26695 were treated by alkylation and reduction cocktail, and then fractionated on thiol-affinity chromatography. The protein mixture eluted from thiol-affinity chromatography was analyzed by resolving on 12% SDS-PAGE and staining with coomassie blue (Fig. 1). Major bands of whole cell extract were reduced, and several bands newly appeared on the eluted fraction as shown in Fig. 1.
The eluted fraction of H. pylori strain 26695 on the thiol-affinity chromatography was treated with 2-DE clean up kit for removing contaminants such as, SDS, DTT, etc. The purified sample by cleaning kit was loaded onto the precast IPG strips (17 cm) of a pH gradient ranging from 5.0~8.0 and 6.0~11.0 for the 1-D protein isoelectric focusing. The strips were loaded onto a 13% acrylamide gel of 18 × 20 cm for 2-D electrophoresis. After running SDS-PAGE, the separated spots were visualized by silver staining (Fig. 2). Represented protein spots on 2-DE gels were identified by MALDI-TOF-MS (Table 1). In this study, 67 spots were identified, which represented 35 genes. Several proteins, such as chaperone and heat shock protein (GroEL, HP0010), translation elongation factor EF-G (FusA, HP1195), translation elongation factor EF-Tu (FusB, HP1205), alkylhydroperoxide reductase (AhpC, HP1563), and thioredoxin (TrxA, HP0824) showed a high abundance on the 2-DE gel, where alkylhydroperoxide reductase (AhpC, HP1563) was the most abundant protein.
It is possible that protective factors against environmental oxidative stresses are sited in the periplasmic space of H. pylori. To identify thiol-active protein in periplasmic space of H. pylori, they were treated with hypertonic solution and suspended with cold distilled water. The periplasmic fraction was the separated from the pellet by centrifugation. The periplasmic fraction and the pellet were then subjected to 2-DE to compare their protein spot profiles. As shown in Fig. 3, the spot profiles of 2-DE were similar to each other. Among 93 protein spots definitely stained in 2-DE, 22 spots showed more than 2-fold higher intensity in the periplasmic fraction than in the pellet when their intensities were determined by PDQUEST program. Of them, 11 protein spots could be identified by peptide fingerprint using MALD-TOF MS, as shown Table 2. Here, 4 proteins such as HypB (HP0900), FusA (HP1195), TufB (HP1205), and AhpC (HP1563) were thiol-active proteins.
Cysteine is one of the most rarely used amino acids in the proteins of most organisms studied so far. Therefore, when highly conserved in proteins, it usually plays crucial roles in the structure, function, or regulation of the protein (16, 17). The redox regulation of protein function plays an important role in many biological processes. Here, comprehensive analysis of disulfide-containing proteins of H. pylori whole cell extract was attempted, using thiol-affinity chromatography and proteome analysis. In this study, total proteins extracted from H. pylori strain 26695 were treated with thiol-alkylating cocktail, containing both N-ethylmaleimide and iodoacetamide for blocking the free thiol residues of proteins. After complete removal of alkylating reagents, disulfide bonds were reduced to produce free thiols with tributylphosphine. Reduced proteins with one or more free thiol groups were covalently captured on a thiol-sepharose column. Thiol groups of proteins were covalently bound to the resin via a thiol-disulfide exchange reaction. This chromatography was developed to purify proteins that have reactive thiol groups. However, thiol-exchange reaction of resin was exploited to isolate disulfide-containing proteins through alkylation and reduction of whole cell extract. In this way, disulfide containing proteins could be specifically captured and separated from H. pylori whole cell extracts.
Treatment of SDS was inevitable for alkylation and reduction of proteins in the whole cell extract. In several previously studies, 2-DE resolution was not satisfactory for proteomic analysis when treated with SDS during sample preparation (58, 59). This problem may have been because of the incomplete removal of SDS, and other charged materials, interfering with isoelectric focusing. Hence, we applied extensive sample cleaning procedures in order to completely remove contaminants, using TCA precipitation and a commercial sample cleaning kit, resulting in protein samples which were clear enough to be displayed on 2-DE (Fig. 2).
In this study, 35 disulfide-containing proteins were identified in the protein mixture eluted through thiol-affinity chromatography. These proteins were related with post-translational modification, energy production, metabolism, and hypothetical proteins. AhpC (HP1563) showed the highest intensity on the 2-DE gel of the eluted fraction of thiol-affinity chromatography, H. pylori AhpC is known to be an essential protein for protection against oxidative damage, and to have a disulfide bond (60, 61). Trx-1 (HP0824) has previously been shown to be an electron donor in vitro for AhpC (62), and to behave as a stress response element as broth grown bacteria secreted Trx in response to chemical, biological, and environmental stresses (63). H. pylori GroES (HP0011) has 6 cysteine residues, contains a unique His-rich C-terminal extension, and was demonstrated to bind nickel in vitro (64). Other pathogenic bacteria did not contain cysteine residues. GroEL (HP0010) forms heptameric barrel complex with GroES, which mediated the correct folding of a variety of cellular proteins and which is highly conserved and essential in all living organisms (65). GroEL was abundantly observed in the eluted fraction of thiol-affinity chromatography even though it contained one cycteine residue, possibly due to the incomplete dissociation of GroEL-GroES hexameric complexes during sample preparation. UreB (urease beta subunit) is known to the most abundant protein in the whole cell extract, but the intensity of the protein spot from eluted fraction of thiol-affinity chromatography was reduced on 2-DE gel. It seemed to be contaminated during the procedure of disulfide containing proteins separation. The structural study demonstrated that H. pylori urease does not have disulfide bonds and subunits interaction of this enzyme are ionic and based on hydrogen bonds (66, 67). Three secreted proteins were identified, two hypothetical cysteine rich proteins (HP0211, HP0235) and Msr (Peptide methionine sulfoxide reductase). HP0211 has a disulfide bond and β-lactamase activity (68), HP0235 was reported to be one of the slr gene family (69) and Msr has an important function as a repair enzyme for proteins that have been inactivated by oxidation (70). Other identified proteins were related to protein biosynthesis (HP0124, HP0476), amino acid biosynthesis (HP0566, HP0663), lipopolysaccharide biosynthesis (HP0857, HP0858), translation (HP1195, HP1205), and chaperone (HP0010, HP0011, and HP0109) etc., as well as unknown functions (HP1588). A number of investigations have demonstrated the existence of complex oxidative-reductive pathways in the bacterial periplasm (71, 72). In this study, disulfide-containing proteins of HypB (HP0900), FusA (HP1195), TufB (HP1205), and AhpC (HP1563) showed enhanced intensities in the periplasmic space (Fig. 3 and Table 2), demonstrating that these proteins might be located in the outer region of the plasmic membrane and act as first defense factors against environmental oxidative stresses.
Finally, this study suggests that thiol-affinity chromatography is an effective tool of separation and identification using the 2-DE system and mass spectrometry of disulfide containing proteins from total cell extracts. In particular, 17 of the identified proteins were newly identified for H. pylori. Disulfide containing proteins identified in this study will provide the standard landscape for constructing the proteome components responsible for redox regulation of H. pylori.
Figures and Tables
Figure 1
SDS-PAGE analysis of flow-through and eluted fraction in the thiol-affinity chromatography applied to H. pylori 26695 whole cell extract that was subjected to alkylation and reduction. M, protein size marker; 1, the whole cell extract; 2, thiol-affinity-unbound protein, flow-through fraction; 3, thiol-affinity-bound proteins, eluted fraction. Proteins were resolved with 12% SDS-gels. The gel was stained with coomassie blue.
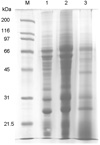
Figure 2
Two-dimensional gel electrophoresis profiles of the whole cell extract (A and C) and the eluted fraction of thiol-affinity chromatography (B and D) of H. pylori 26695. The protein mixture (200 µg) were separated on IPG strip of pH 5.0~8.0 and 6.0~11.0 by 13% SDS-PAGE and then visualized by silver staining. The spot numbers on the gels are given in Tigr locus name of Table 1.
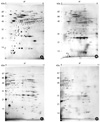
Figure 3
Two-dimensional gel electrophoresis of periplasmic fraction (100 µg) of H. pylori strain 26695. The proteins were separated on an immobilized pH gradient strip and subsequently on a 13% SDS-PAGE gel. The gels were stained by silver staining and were scanned and processed using Fluor-S MutiImager system and the PDQUEST-2D Gel analysis software (Bio-rad, vesion 7.01, CA, USA). Locus No. represent protein spots showed >2-fold higher intensity in the periplasmic fraction (B) than in the pellet (A).

Table 1
Putative disulfide containing proteins identified by MALDI-TOF-MS

a Locus numbers are marked in Fig. 2.
b Number of cysteine residues
c C, Energy production and conversion; E, Amino acid transport and metabolism; H, Coenzyme transport and metabolism; J, Translation, ribosomal structure and biogenesis; K, Transcription; M, Cell wall/membrane/envelope biogenesis; O, Posttranslational modification, protein turnover, chaperones; R, General function prediction only; S, Uncharacterized protein conserved in bacteria; T, Signal transduction mechanisms.
d Cellular functions are according to NCBI and TIGR database.
References
1. Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984. 1:1311–1315.

2. Graham DY, Malaty HM, Evans DG, Evans DJ Jr, Klein PD, Adam E. Epidemiology of Helicobacter pylori in an asymptomatic population in the United States. Effect of age, race, and socioeconomic status. Gastroenterology. 1991. 100:1495–1501.

3. Wolle K, Leodolter A, Malfertheiner P, König W. Antibiotic susceptibility of Helicobacter pylori in Germany: stable primary resistance from 1995 to 2000. J Med Microbiol. 2002. 51:705–709.

4. Aktay AN, Werlin SL, Hellman RS. The impact of solid-phase gastric emptying studies in the management of children with dyspepsia. Clin Pediatr (Phila). 2003. 42:621–625.

5. Dooley CP, Cohen H, Fitzgibbons PL, Bauer M, Appleman MD, Perez-Perez GI, et al. Prevalence of Helicobacter pylori infection and histologic gastritis in asymptomatic persons. N Engl J Med. 1989. 321:1562–1566.

6. Blaser MJ. Gastric Campylobacter-like organisms, gastritis, and peptic ulcer disease. Gastroenterology. 1987. 93:371–383.

7. Sidebotham RL, Baron JH. Hypothesis: Helicobacter pylori, urease, mucus, and gastric ulcer. Lancet. 1990. 335:193–195.
8. Mégraud F, Lamouliatte H. Helicobacter pylori and duodenal ulcer. Evidence suggesting causation. Dig Dis Sci. 1992. 37:769–772.
9. Baik SC, Youn HS, Chung MH, Lee WK, Cho MJ, Ko GH, et al. Increased oxidative DNA damage in Helicobacter pylori-infected human gastric mucosa. Cancer Res. 1996. 56:1279–1282.
10. Forman D, Newell DG, Fullerton F, Yarnell JW, Stacey AR, Wald N, et al. Association between infection with Helicobacter pylori and risk of gastric cancer: evidence from a prospective investigation. BMJ. 1991. 302:1302–1305.

11. Nomura A, Stemmermann GN, Chyou PH, Kato I, Perez-Perez GI, Blaser MJ. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991. 325:1132–1136.

12. Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991. 325:1127–1131.

13. Mendz GL, Jimenez BM, Hazell SL, Gero AM, O'Sullivan WJ. De novo synthesis of pyrimidine nucleotides by Helicobacter pylori. J Appl Bacteriol. 1994. 77:1–8.
14. Burns BP, Hazell SL, Mendz GL. Acetyl-CoA carboxylase activity in Helicobacter pylori and the requirement of increased CO2 for growth. Microbiology. 1995. 141:3113–3118.

15. Joo JS, Park KC, Song JY, Kim DH, Lee KJ, Kwon YC, et al. A thin-layer liquid culture technique for the growth of Helicobacter pylori. Helicobacter. 2010. 15:295–302.

16. Kettenhofen NJ, Wood MJ. Formation, reactivity, and detection of protein sulfenic acids. Chem Res Toxicol. 2010. 23:1633–1646.

17. Leonard SE, Carroll KS. Chemical 'omics' approaches for understanding protein cysteine oxidation in biology. Curr Opin Chem Biol. 2011. 15:88–102.

18. Klomsiri C, Karplus PA, Poole LB. Cysteine-based redox switches in enzymes. Antioxid Redox Signal. 2011. 14:1065–1077.

19. Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol. 2007. 7:381–391.

20. Sarajärvi T, Tuusa JT, Haapasalo A, Lackman JJ, Sormunen R, Helisalmi S, et al. Cysteine 27 variant of the delta-opioid receptor affects amyloid precursor protein processing through altered endocytic trafficking. Mol Cell Biol. 2011. 31:2326–2340.

21. Subramanian S, Sijwali PS, Rosenthal PJ. Falcipain cysteine proteases require bipartite motifs for trafficking to the Plasmodium falciparum food vacuole. J Biol Chem. 2007. 282:24961–24969.

22. Guttmann RP. Redox regulation of cysteine-dependent enzymes. J Anim Sci. 2010. 88:1297–1306.
23. Spadaro D, Yun BW, Spoel SH, Chu C, Wang YQ, Loake GJ. The redox switch : dynamic regulation of protein function by cysteine modifications. Physiol Plant. 2010. 138:360–371.

24. Cross JV, Templeton DJ. Regulation of signal transduction through protein cysteine oxidation. Antioxid Redox Signal. 2006. 8:1819–1827.

25. Franklin CC, Backos DS, Mohar I, White CC, Forman HJ, Kavanagh TJ. Structure, function, and post-translational regulation of the catalytic and modifier subunits of glutamate cysteine ligase. Mol Aspects Med. 2009. 30:86–98.

26. Stipanuk MH, Ueki I, Dominy JE Jr, Simmons CR, Hirschberger LL. Cysteine dioxygenase: a robust system for regulation of cellular cysteine levels. Amino Acids. 2009. 37:55–63.

27. Turell L, Botti H, Carballal S, Radi R, Alvarez B. Sulfenic acid--a key intermediate in albumin thiol oxidation. J Chromatogr B Analyt Technol Biomed Life Sci. 2009. 877:3384–3392.

28. Bardwell JC, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991. 67:581–589.

29. Gleiter S, Bardwell JC. Disulfide bond isomerization in prokaryotes. Biochim Biophys Acta. 2008. 1783:530–534.

30. Sevier CS, Kaiser CA. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta. 2008. 1783:549–556.

31. Inaba K. Structural basis of protein disulfide bond generation in the cell. Genes Cells. 2010. 15:935–943.
32. Gruber CW, Cemazar M, Heras B, Martin JL, Craik DJ. Protein disulfide isomerase: the structure of oxidative folding. Trends Biochem Sci. 2006. 31:455–464.

33. Okazaki S, Tachibana T, Naganuma A, Mano N, Kuge S. Multistep disulfide bond formation in Yap1 is required for sensing and transduction of H2O2 stress signal. Mol Cell. 2007. 27:675–688.

34. O'Brian CA, Chu F. Post-translational disulfide modifications in cell signaling--role of inter-protein, intra-protein, S-glutathionyl, and S-cysteaminyl disulfide modifications in signal transmission. Free Radic Res. 2005. 39:471–480.
35. Hansen RE, Winther JR. An introduction to methods for analyzing thiols and disulfides: Reactions, reagents, and practical considerations. Anal Biochem. 2009. 394:147–158.

36. Lindahl M, Mata-Cabana A, Kieselbach T. The disulfide proteome and other reactive cysteine proteomes: analysis and functional significance. Antioxid Redox Signal. 2011. 14:2581–2642.

37. Kumar JK, Tabor S, Richardson CC. Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc Natl Acad Sci U S A. 2004. 101:3759–3764.

38. Pérez-Pérez ME, Florencio FJ, Lindahl M. Selecting thioredoxins for disulphide proteomics: target proteomes of three thioredoxins from the cyanobacterium Synechocystis sp. PCC 6803. Proteomics. 2006. 6:S186–S195.
39. Hochgräfe F, Mostertz J, Albrecht D, Hecker M. Fluorescence thiol modification assay: oxidatively modified proteins in Bacillus subtilis. Mol Microbiol. 2005. 58:409–425.

40. Leichert LI, Jakob U. Protein thiol modifications visualized in vivo. PLoS Biol. 2004. 2:e333.
41. Hu W, Tedesco S, McDonagh B, Bárcena JA, Keane C, Sheehan D. Selection of thiol- and disulfide-containing proteins of Escherichia coli on activated thiol-sepharose. Anal Biochem. 2010. 398:245–253.

42. Brandes N, Rinck A, Leichert LI, Jakob U. Nitrosative stress treatment of E. coli targets distinct set of thiol-containing proteins. Mol Microbiol. 2007. 66:901–914.

43. Charles RL, Schröder E, May G, Free P, Gaffney PR, Wait R, et al. Protein sulfenation as a redox sensor: proteomics studies using a novel biotinylated dimedone analogue. Mol Cell Proteomics. 2007. 6:1473–1484.
44. Yano H, Kuroda S. Introduction of the disulfide proteome: application of a technique for the analysis of plant storage proteins as well as allergens. J Proteome Res. 2008. 7:3071–3079.

45. Yang Y, Song Y, Loscalzo J. Regulation of the protein disulfide proteome by mitochondria in mammalian cells. Proc Natl Acad Sci U S A. 2007. 104:10813–10817.

46. García-Santamarina S, Boronat S, Espadas G, Ayté J, Molina H, Hidalgo E. The oxidized thiol proteome in fission yeast--optimization of an ICAT-based method to identify H2O2-oxidized proteins. J Proteomics. 2011. 74:2476–2486.

47. McDonagh B, Padilla CA, Pedrajas JR, Bárcena JA. Biosynthetic and iron metabolism is regulated by thiol proteome changes dependent on glutaredoxin-2 and mitochondrial peroxiredoxin-1 in Saccharomyces cerevisiae. J Biol Chem. 2011. 286:15565–15576.

48. Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010. 468:790–795.

49. McDonagh B, Sheehan D. Effect of oxidative stress on protein thiols in the blue mussel Mytilus edulis: proteomic identification of target proteins. Proteomics. 2007. 7:3395–3403.

50. Wang H, Qian WJ, Chin MH, Petyuk VA, Barry RC, Liu T, et al. Characterization of the mouse brain proteome using global proteomic analysis complemented with cysteinyl-peptide enrichment. J Proteome Res. 2006. 5:361–369.

51. Liu T, Qian WJ, Chen WN, Jacobs JM, Moore RJ, Anderson DJ, et al. Improved proteome coverage by using high efficiency cysteinyl peptide enrichment: the human mammary epithelial cell proteome. Proteomics. 2005. 5:1263–1273.

52. Song JY, Choi YJ, Kim JM, Kim YR, Jo JS, Park JS, et al. Purification and characterization of Helicobacter pylori γ-glutamyltranspeptidase. J Bacteriol Virol. 2011. 41:255–265.

53. Lee K, Lee J, Kim Y, Bae D, Kang KY, Yoon SC, et al. Defining the plant disulfide proteome. Electrophoresis. 2004. 25:532–541.

54. Rohde M, Püls J, Buhrdorf R, Fischer W, Haas R. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol. 2003. 49:219–234.

55. Park JW, Lee SG, Song JY, Jun JS, Joo JS, Youn HS, et al. Proteomic analysis of Helicoacter pylori whole cell proteins using the narrow range IPG strips. J Bacteriol Virol. 2007. 37:203–212.

56. Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999. 20:601–605.

57. O'Connell KL, Stults JT. Identification of mouse liver proteins on two-dimensional electrophoresis gels by matrix-assisted laser desorption/ionization mass spectrometry of in situ enzymatic digests. Electrophoresis. 1997. 18:349–359.
58. Harder A, Wildgruber R, Nawrocki A, Fey SJ, Larsen PM, Görg A. Comparison of yeast cell protein solubilization procedures for two-dimensional electrophoresis. Electrophoresis. 1999. 20:826–829.

59. Nandakumar MP, Marten MR. Comparison of lysis methods and preparation protocols for one- and two-dimensional electrophoresis of Aspergillus oryzae intracellular proteins. Electrophoresis. 2002. 23:2216–2222.

60. Wang G, Alamuri P, Maier RJ. The diverse antioxidant systems of Helicobacter pylori. Mol Microbiol. 2006. 61:847–860.

61. Chuang MH, Wu MS, Lo WL, Lin JT, Wong CH, Chiou SH. The antioxidant protein alkylhydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc Natl Acad Sci U S A. 2006. 103:2552–2557.

62. Comtois SL, Gidley MD, Kelly DJ. Role of the thioredoxin system and the thiol-peroxidases Tpx and Bcp in mediating resistance to oxidative and nitrosative stress in Helicobacter pylori. Microbiology. 2003. 149:121–129.

63. Windle HJ, Fox A, Ní Eidhin D, Kelleher D. The thioredoxin system of Helicobacter pylori. J Biol Chem. 2000. 275:5081–5089.
64. Schauer K, Muller C, Carrière M, Labigne A, Cavazza C, De Reuse H. The Helicobacter pylori GroES cochaperonin HspA functions as a specialized nickel chaperone and sequestration protein through its unique C-terminal extension. J Bacteriol. 2010. 192:1231–1237.

66. Ha NC, Oh ST, Sung JY, Cha KA, Lee MH, Oh BH. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nat Struct Biol. 2001. 8:505–509.
67. Pinkse MW, Maier CS, Kim JI, Oh BH, Heck AJ. Macromolecular assembly of Helicobacter pylori urease investigated by mass spectrometry. J Mass Spectrom. 2003. 38:315–320.

68. Mittl PR, Lüthy L, Hunziker P, Grütter MG. The cysteine-rich protein A from Helicobacter pylori is a beta-lactamase. J Biol Chem. 2000. 275:17693–17699.
69. Ogura M, Perez JC, Mittl PR, Lee HK, Dailide G, Tan S, et al. Helicobacter pylori evolution: lineage-specific adaptations in homologs of eukaryotic Sel1-like genes. PLoS Comput Biol. 2007. 3:e151.
70. Alamuri P, Maier RJ. Methionine sulphoxide reductase is an important antioxidant enzyme in the gastric pathogen Helicobacter pylori. Mol Microbiol. 2004. 53:1397–1406.

71. Sardesai AA, Genevaux P, Schwager F, Ang D, Georgopoulos C. The OmpL porin does not modulate redox potential in the periplasmic space of Escherichia coli. EMBO J. 2003. 22:1461–1466.

72. Genevaux P, Bauda P, DuBow MS, Oudega B. Identification of Tn10 insertions in the dsbA gene affecting Escherichia coli biofilm formation. FEMS Microbiol Lett. 1999. 173:403–409.

73. Cho MJ, Jeon BS, Park JW, Jung TS, Song JY, Lee WK, et al. Identifying the major proteome components of Helicobacter pylori strain 26695. Electrophoresis. 2002. 23:1161–1173.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download