Abstract
A total of 91 non-typhoid Salmonella isolated from pediatric patients with diarrhea in Seoul from 2003 to 2009 was tested for antimicrobial susceptibility of nalidixic acid (NA). Forty strains of NA resistance or intermediate susceptible non-typhoid Salmonella were identified and their minimum inhibitory concentrations (MICs) of NA, ciprofloxacin (CIP), and norfloxacin (NOR) were determined. Of the 40 isolates, 26 were resistant to NA (MIC >256 µg/ml). Only one isolate was high-level resistant to CIP (12 µg/ml) and NOR (48 µg/ml). Mutations in gyrA and parC genes were studied by PCR and sequencing. All NA-resistant isolates carried point mutations in the gyrA quinolone resistance determining regions (QRDR) at codon 83 or 87 (MICs of NA, >256 µg/ml; MICs of CIP, 0.047~0.25 µg/ml; MICs of NOR, 0.38~1.5 µg/ml). A double change in GyrA was found in one Salmonella Enteritidis (MIC of CIP, 12 µg/ml; MIC of NOR, 48 µg/ml). In respect of the ParC protein, a single change at Thr57→Ser was found in 3 isolates (MICs of NA, >256 µg/ml; MICs of CIP, 0.19~0.25 µg/ml; MICs of NOR, 1 µg/ml). At the same time, these strains changed from Ser83 to Tyr in the gyrA. The result of the investigation for the prevalence of plasmid-mediated quinolone resistance (PMQR) genes, 14 isolates harbored qnr gene among 40 isolates. All of 14 isolates showed decreased susceptibility at NA (MICs 4~16 µg/ml) and except one strain, all of qnr genes were identified as qnrB. Mutations in the gyrA gene and production of PMQR determinants were critical for quinolone resistance and decreased susceptibility to fluoroquinolone in these isolates.
Salmonella enterica is widespread in humans and animals worldwide and is of increasing public health concern as causative pathogens of food poisoning (1). While approximately 2000 serotypes of Salmonella have been associated with enterocolitis, Salmonella Typhimurium and Salmonella Enteritidis are two major etiologic agents of food-borne salmonellosis in human (2). Non-typhoidal salmonellosis in humans is usually a self-limiting disease confined to the gastrointestinal tract, therefore antibiotic treatment is not recommended. However, antibiotic therapy may be required in immunocopromised patients, in cases of extraintestinal infections, and for infants and elderly peoples (3). In the treatment of the non-typhoid Salmonella, the ciprofloxacin is used and recently it is the tendency that the resistance is gradually increased to the all, globally, the quinolone antibiotic (4, 5). In 2011, according to Korea Center for Disease Control and Prevention (KCDC) report, the nalidixic acid (NA) resistance rate in clinical Salmonella isolates was 4.8% in 1999 and increased to 62.1% in 2009.
The first quinolone, NA, was introduced into clinical use at the beginning of the 1960s (6). In the 1980s, ciprofloxacin (CIP), a fluoroquinolone with a wide spectrum of in vitro antibacterial activity, particularly against gram negative bacteria, first became clinically available (7). However, this high level of use, and to some degree of misuse in the sense of unnecessary use, or use of quinolones with poor activity in some developing countries, has been blamed for the rapid development of bacterial resistance to these agents (4).
Quinolone resistance in Gram-negative bacteria is primarily attributable to mutations in the quinolone resistance determining regions (QRDR) consisting of the gyrA and parC genes, which are the subunits of the target enzymes of quinolones, DNA gyrase subunit A and topoisomerase IV, respectively (8). Other mechanisms of resistance, such as efflux pump systems or modifications of porins, can decrease susceptibility to quinolone (9). In addition, decreased susceptibility to quinolones can be mediated by plasmid-mediated quinolone resistance genes, named qnr, aac(6')-Ib-cr and qepA (7).
Research on the mechanisms of resistance to quinolones of Salmonella at the early 2000's was reported by several authors in Korea, but there are few reports on the latest strains. In the present study, we investigated the mutations in the QRDR and the prevalence of PMQR determinants of NA resistance or intermediate Salmonella isolated from pediatric patients with diarrhea in order to figure out their distribution and significance.
From 2003 to 2009, a total 91 non-typhoid Salmonella strains were isolated from pediatric patients with diarrhea at the Seoul Metropolitan Government Research Institute of Public Health and Environment (SIHE) in Seoul, Korea. After performing the biochemical identification by API 20E Kit (BioMerieux Co. Mo, USA) about K/A and H2S formation cell group among the colony growing in MacConkey agar plate into the colorless, the Salmonella serotypes were determined by slide agglutination according to the Kauffmann-White scheme using O and H-antisera (Difco Co., Detroit, USA). All isolates were stored at -70℃.
NA susceptibility of Salmonella isolates was determined using a disc diffusion method, according to the guidelines of the Clinical Laboratories Standards Institute (CLSI). Determination of MIC was performed using E-tests (AB Biodisk, Solna, Sweden) on Mueller Hinton plates following the manufacturer's recommendations. The antimicrobial agents tested were NA, CIP, and norfloxacin (NOR).
The QRDRs of gyrA and parC genes were amplified by PCR from the DNA of 26 NA-resistant isolates. In brief, colonies were suspended in 200 µl of distilled water and boiled to prepare DNA templates. The primer set used was those described by Giraud et al. (10) (Table 1). The PCR conditions were 3 min at 94℃ and 30 cycles of 94℃ for 1 min, 55℃ for 1 min (parC 52℃ 1 min), and 72℃ for 1 min. Positive PCR products were purified using a PCR purification kit (Cosmo Inc., Seoul, Korea), and DNA sequencing was performed by Cosmo Inc. (Cosmo Inc., Seoul, Korea).
Among 91 isolates, a total of 40 strains of NA resistance or intermediate susceptible non-typhoid Salmonella were obtained from pediatric patients with diarrhea from 2003 to 2009. The most frequently identified serotypes were Salmonella Enteritidis, accounting for 37.5% (15 isolates) followed by Montevideo (27.5%, 11 isolates), Hillingon (12.5%, 5 isolates), and Typhimurium (7.5%, 3 isolates). The remaining 6 isolates belonged to other serotypes.
Antimicrobial susceptibility testing by disc method revealed that 28 isolates were resistant to NA, whereas 12 isolates were intermediate susceptible to NA (Table 2). Yearly, the 1 of 11 isolates (9.1%) in 2003, 1 of 9 isolates (11.1%) in 2004, 1 of 11 isolates (9.1%) in 2005, 9 of 26 isolates (34.6%) in 2006, 6 of 18 isolates (33.3%) in 2007, 5 of 9 isolates (55.6%) in 2008, 5 of 7 isolates (71.4%) in 2009 was shown resistant to NA by disc test. The rate of resistance has been increasing since 2006 (Table 2). The MIC values of NA resistance and intermediate susceptible Salmonella isolates are shown in Table 2. Of the 28 isolates, 26 were resistant to NA (MIC >256 µg/ml) by MIC test. One Salmonella Enteritidis (No. 30) isolated in 2007 exhibited a high level of resistance to both CIP (12 µg/ml) and NOR (48 µg/ml). The remaining isolates for CIP and NOR were 0.047~0.25 µg/ml and 0.25~2 µg/ml, respectively. Among the 40 isolates, 19 (47.5%) showed increased MIC (≥ 0.125 µg/ml) for CIP.
PCR of gyrA and parC genes was performed on a total of 26 NA resistant isolates and DNA sequencing of the PCR products was carried out. A single amino acid substitution in the QRDR of GyrA protein was detected in all isolates (MIC >256 µg/ml) (Table 3). All mutations occurred in the Ser83 and Asp87 codon, and the variation observed much in the codon 87. The substitutions were as follows (number of strains): Ser83 → Tyr (3), Ser83 → Leu (2), Ser83 → Phe (1), Asp87 → Asn (11), Asp87 → Gly (7), and Asp87 → Tyr (2). One Salmonella Enteritidis isolated in 2007 (No. 30) had the double mutations in gyrA Ser83 → Leu and Asp87 → Tyr. In respect of the ParC protein, a single change at Thr57 → Ser was found in 3 isolates (MICs of NA, >256 µg/ml; MICs of CIP, 0.19~0.25 µg/ml; MICs of NOR, 1 µg/ml). At the same time, these strains changed from Ser83 to Tyr in the gyrA.
The PMQR genes were detected in 14 (35%) of the 40 Salmonella isolates (Table 3). The most commonly observed qnr-positive serotypes were Montevideo (N=11), followed by Othmarschen (N=2), and Typhimurium (N=1). The qnrB gene was detected in 13 of the 14 qnr-positive isolates and the MICs of NA ranged from 4 µg/ml to 16 µg/ml. The qnrA gene and aac(6')-Ib-cr were found in only 1 strain and MICs of NA, CIP, and NOR were 8 µg/ml, 0.25 µg/ml, and 2 µg/ml, respectively. The qnrS and qepA genes were not detected in any of the Salmonella strains in this study.
In the current study, we investigated mechanisms of quinolone resistance of NA-resistance or intermediate susceptibility Salmonella isolated from pediatric patients with diarrhea in order to figure out their prevalence and characterization. In Korea, the NA resistance rate in clinical Salmonella isolates was 1.8% in 1995-1996, but has increased to 21.8% in 2000-2002 (5). In this study, we found that the resistance rate had increased gradually from 9~11% in 2003-2005 to 34.6~71.4% in 2006-2009. In spite of being the strains isolated from the child, the resistance rate of the NA was increased over time and the injury reached a serious level.
The results of MICs for quinolone and fluoroquinolone were 26 isolates (28.5%) with high resistance to NA between 2003 and 2009. Except one, all isolates were 0.064~0.25 µg/ml and 0.25~2 µg/ml in MICs for CIP and NOR, respectively, and this means that the resistance of fluoroquinolones is still uncommon. The results are similar to those in previous reports (14~16). Lee et al. (17) in Korea has reported that fluoroquinolone resistance of Salmonella Haardt isolated from chicken was a little higher than for the humans.
Consistent with previous reports, we found that the main mechanism of NA resistance was single point mutations in the QRDR region of the gyrA gene at codons 83 or 87. In this study, Ser83 changed at Tyr, Leu, and Phe and Asp87 changed at Asn, Gly, and Tyr. These results were same with the substitution which had been reported as the most frequent substitution in GyrA of Salmonella (5, 14, 16, 18, 19). A previous study by Fabrega et al. (14) suggested that the position of the mutations in the gyrA gene and the substituting amino acid can be depended on the serovar analysis. In this study, there was the difference in several papers (15, 17) and this reason is considered to be the origin and the serotypes. The occasion mutation of ParC had been reported. Also, the substitution from Thr57 to Ser had been reported as the occasion mutation in ParC (14). In this study, three isolates (two of Haardt, one of Bovismorbificans) showed same substitution form with the substitution from Thr57 to Ser. In Korea, the research of substitution form had been reported by Jeong et al. (19) and Kim et al. (20) and it showed that the substitution from Thr57 to Ser happens occasionally. In Salmonella, given the fact that a single mutation of gyrA gene might cause quinolone resistance and reduced fluoroquinolone susceptibility (21), it was interesting that a double mutation of the gyrA gene happened in the parC. However, there was a discrepancy between Eaves et al.'s (22) and Ling et al.'s (23) research. Eaves et al. suggested that Thr57-Ser substitution in ParC made isolates more sensitive to CIP and formed a hypothesis that the mutation at codon 57 of parC could be a naturally occurring compensatory mutation. In this study, we had the same view on the substitution with Eaves et al.'s research due to our result in which the substitution did not cause an increase of MICs of CIP in ParC.
PMQR mechanisms, including qnr genes, aac(6')-Ib-cr, and qepA, were found to confer decreased susceptibility to fluoroquinolones (24). In this study, the qnr genes had been detected in 14 isolates (35%) that all of the strains showed the decreased susceptibility to NA (MICs 4~16 µg/ml). In particular, one isolate (S. Typhimurium) in which the qnrA and aac(6')-Ib-cr is detected, showed the reduced susceptibility to the quinolone and fluoroquinolones. It could be confirmed to contribute to reduced susceptibility as the production of PMQR determinants. The PMQR genes detected in Salmonella are rarely found in Korea (5, 20). Recently, in Korea, Jeong et al. (19) reported the PMQR detection in Salmonella. However, there was a distinct from our study in that Jeong et al. detected 6 qnr-positive clinical isolates among 284 samples (2.1%) and these had high level resistance to NA. This study was limited to pediatric patients with diarrhea in Seoul and it is interesting that the qnr gene rate is higher. Moreover, this is the first report of the qnrA gene in Salmonella from Korea. The qnr types of the present study correspond well with those of the earlier study (25) which reported that qnr-positive Enterobacteriaceae is had qnrB. In China (26), USA (27), and Japan (28), there were reported aac(6')-Ib-cr and qnr genes detected in Salmonella. Among those researches, there were differences in the prevalence and variant types of PMQR genes. We are now unsure whether the reason is serotypes or geographic region. We will conduct further research on the reason for the difference.
Due to the continuous increase in quinolone resistance of non-typhoid Salmonella in Korea, continued resistance research monitoring is under consideration.
Figures and Tables
Table 2
Antimicrobial susceptibility against nalidixic acid of 91 nontyphoid Salmonella isolated in Seoul from 2003 to 2009

Table 3
The minimum inhibitory concentration and quinolone resistance mechanisms of 40 nontyphoid-Salmonella isolates
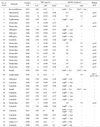
aA total of 26 nalidixic acid- resistant isolates were examined
bNA, nalidixic acid; MIC value: susceptible (≤16 µg/ml), intermediate (-), resistance (≥32 µg/ml)
cCIP, ciprofloxacin; MIC value: susceptible (≤1 µg/ml), intermediate (2 µg/ml), resistance (≥4 µg/ml)
dNOR, norfloxacin; MIC value: susceptible (≤4 µg/ml), intermediate (8 µg/ml), resistance (≥16 µg/ml)
eNT indicate not tested
f(-) indicate no mutation or no PMQR genes.
gThe variant of the S. Typhimurium, presently, the serotype is not named.
References
1. Collazo CM, Galán JE. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella Typhimurium. Infect Immun. 1996. 64:3524–3531.

2. Darwin KH, Miller VL. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin Microbiol Rev. 1999. 12:405–428.

3. Nógrády N, Gadó I, Tóth A, Pászti J. Antibiotic resistance and class 1 integron patterns of non-typhoidal human Salmonella serotypes isolated in Hungary in 2002 and 2003. Int J Antimicrob Agents. 2005. 26:126–132.

4. Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003. 51:1109–1117.

5. Choi SH, Woo JH, Lee JE, Park SJ, Choo EJ, Kwak YG, et al. Increasing incidence of quinolone resistance in human non-typhoid Salmonella enterica isolates in Korea and mechanisms involved in quinolone resistance. J Antimicrob Chemother. 2005. 56:1111–1114.

6. Ball P. Quinolone generations: natural history or natural selection? J Antimicrob Chemother. 2000. 46:17–24.

7. Paton JH, Reeves DS. Fluoroquinolone antibiotics. Microbioloy, pharmacokinetics and clinical use. Drugs. 1988. 36:193–228.
8. Vila J. White DG, Alekshun MN, McDermotte PF, editors. Fluoroquinolone resistance. Frontiers in antimicrobial resistance: a tribute to Stuart B. Levy. 2005. Herndon, VA: ASM Press;41–52.

9. Everett MJ, Jin YF, Ricci V, Piddock LJ. Contributions of individual mechanisms to fluoroquinolone resistance of 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother. 1996. 40:2380–2386.

10. Giraud E, Brisabois A, Martel JL, Chaslus-Dancla E. Comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone resistant strains in the field. Antimicrob Agents Chemother. 1999. 43:2131–2137.

11. Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC. Qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother. 2006. 50:2872–2874.

12. Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. Prevalence in the United States of aac(6')-Ib-cr encoding a ciprofloxacin modifying enzyme. Antimicrob Agents Chemother. 2006. 50:3953–3955.

13. Kang HY, Tamang MD, Seol SY, Kim JM. Dissemination of plasmid-mediated qnr, aac(6)-Ib-cr, and qepA genes among 16S rRNA methylase producing Enterobacteriaceae in Korea. J Bacteriol Virol. 2009. 39:173–182.

14. Fàbrega A, Sánchez-Céspedes J, Soto S, Vila J. Quinolone resistance in the food chain. Int J Antimicrob Agents. 2008. 31:307–315.

15. Hwang IS, Song JY, Kim WJ, Jeong HW, Kim MS, Cheong HJ. Prevalence and mechanisms of low level quinolone resistance among non-typhoidal Salmonella isolates from human and poultry/livestock in Korea: usefulness of nalidixic acid resistance test. Infect Chemother. 2010. 42:230–236.

16. Lunn AD, Fàbrega A, Sánchez-Céspedes J, Vila J. Prevalence of mechanisms decreasing quinolone-susceptibility among Salmonella spp. clinical isolates. Int Microbiol. 2010. 13:15–20.
17. Lee K, Lee M, Lim J, Jung J, Park Y, Lee Y. Contamination of chicken meat with Salmonella enterica serovar Haardt with nalidixic acid resistance and reduced fluoroquinolone susceptibility. J Microbiol Biotechnol. 2008. 18:1853–1857.
18. Hamidian M, Tajbakhsh M, Tohidpour A, Rahbar M, Zali MR, Walther-Rasmussen J. Detection of novel gyrA mutations in nalidixic acid-resistant isolates of Salmonella enterica from patients with diarrhoea. Int J Antimicrob Agents. 2011. 37:360–364.

19. Jeong HS, Kim JA, Shin JH, Chang CL, Jeong J, Cho JH, et al. Prevalence of plasmid-mediated quinolone resistance and mutations in the gyrase and topoisomerase IV genes in Salmonella isolated from 12 tertiary-care hospitals in Korea. Microb Drug Resist. 2011. 17:551–557.

20. Kim KY, Park JH, Kwak HS, Woo GJ. Characterization of the quinolone resistance mechanism in foodborne Salmonella isolates with high nalidixic acid resistance. Int J Food Microbiol. 2011. 146:52–56.

21. Piddock LJ. Mechanisms of fluoroquinolone resistance: an update 1994-1998. Drugs. 1999. 58:Suppl 2. 11–18.

22. Eaves DJ, Randall L, Gray DT, Buckley A, Woodward MJ, White AP, et al. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob Agents Chemother. 2004. 48:4012–4015.

23. Ling JM, Chan EW, Lam AW, Cheng AF. Mutations in topoisomerase genes of fluoroquinolone-resistant Salmonellae in Hong Kong. Antimicrob Agents Chemother. 2003. 47:3567–3573.

24. Cattoir V, Nordmann P. Plasmid-mediated quinolone resistance in Gram-nagative bacterial species: an update. Curr Med Chem. 2009. 16:1028–1046.

25. Shin JH, Jung HJ, Lee JY, Kim HR, Lee JN, Chang CL. High rates of plasmid-mediated quinolone resistance QnrB variants among ciprofloxacin-resistant Escherichia coli and Klebsiella pneumoniae from urinary tract infections in Korea. Microb Drug Resist. 2008. 14:221–226.

26. Yu F, Chen Q, Yu X, Pan J, Li Q, Yang L, et al. High prevalence of plasmid-mediated quinolone resistance determinant aac(6')-Ib-cr amongst Salmonella enterica serotype Typhimurium isolates from hospitalised paediatric patients with diarrhoea in China. Int J Antimicrob Agent. 2011. 37:152–155.

27. Sjölund-Karlsson M, Folster JP, Pecic G, Joyce K, Medalla F, Rickert R, et al. Emergence of plasmid-mediated quinolone resistance among non-typhi Salmonella enterica isolates from humans in the United States. Antimicrob Agents Chemother. 2009. 53:2142–2144.

28. Taguchi M, Kawahara R, Seto K, Inoue K, Hayashi A, Yamagata N, et al. Plasmid-mediated quinolone resistance in Salmonella isolated from patients with overseas travelers' diarrhea in Japan. Jpn J Infect Dis. 2009. 62:312–314.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download