Abstract
DNA replication of human cytomegalovirus (HCMV) is a highly regulated process that requires specific interactions between cis-acting lytic origin of replication (oriLyt) and trans-acting viral proteins. Formation of the replication initiation complex is also regulated by specific interactions among viral replication proteins. HCMV replication proteins include origin-binding proteins, core proteins that work in replication forks, and regulatory proteins that modulate host cell functions. This letter describes intriguing questions regarding how HCMV origin-binding proteins interact with oriLyt to initiate DNA replication and how the regulatory UL112-113 proteins, which are found only in beta-herpesviruses, function to promote viral DNA replication.
Figures and Tables
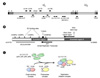
Figure 1
HCMV-encoded replication proteins, the structure of oriLyt, and a possible role of UL112-113 proteins in the formation of the replication initiation complex. (A) The HCMV genome consisting of unique long (UL) and unique short (US) sequences bound by terminal repeats (a/a', b/b', and c/c') and the position of open reading frames (ORFs) for replication proteins are shown. (B) Schematic of HCMV oriLyt showing the relative position of two essential regions, oriLyt promoter, small replicator transcript (SRT), binding sites for C/EBPα and IE2, a highly pyrimidine-rich sequence (Y-block) and the RNA stem-loop structure. Nucleotide (nt) numbers shown are with reference to the AD169 strain DNA sequence. (C) A possible role of UL112-113 proteins as a mediator between the origin-binding complex and the polymerase complex in the formation of the replication initiation complex.
References
1. Mocarski ES, Shenk T, Pass RF. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, editors. Cytomegaloviruses. Fields virology. 2007. Philadelphia, PA.: Lippincott Williams & Wilkins;2701–2772.
2. Anders DG, Punturieri SM. Multicomponent origin of cytomegalovirus lytic-phase DNA replication. J Virol. 1991. 65:931–937.

3. Pari GS, Anders DG. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J Virol. 1993. 67:6979–6988.

4. Chee MS, Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990. 154:125–169.

5. Davison AJ, Dolan A, Akter P, Addison C, Dargan DJ, Alcendor DJ, et al. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J Gen Virol. 2003. 84:17–28.

6. Pari GS, Kacica MA, Anders DG. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J Virol. 1993. 67:2575–2582.

7. Smith JA, Pari GS. Expression of human cytomegalovirus UL36 and UL37 genes is required for viral DNA replication. J Virol. 1995. 69:1925–1931.

8. Iskenderian AC, Huang L, Reilly A, Stenberg RM, Anders DG. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J Virol. 1996. 70:383–392.

9. Pari GS. Nuts and bolts of human cytomegalovirus lytic DNA replication. Curr Top Microbiol Immunol. 2008. 325:153–166.

10. Lee HR, Kim DJ, Lee JM, Choi CY, Ahn BY, Hayward GS, et al. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J Virol. 2004. 78:6527–6542.

11. Kim YE, Lee JH, Kim ET, Shin HJ, Gu SY, Seol HS, et al. Human cytomegalovirus infection causes degradation of Sp100 proteins that suppress viral gene expression. J Virol. 2011. 85:11928–11937.

12. Nevels M, Paulus C, Shenk T. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc Natl Acad Sci U S A. 2004. 101:17234–17239.

13. Huh YH, Kim YE, Kim ET, Park JJ, Song MJ, Zhu H, et al. Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J Virol. 2008. 82:10444–10454.

14. Paulus C, Krauss S, Nevels M. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc Natl Acad Sci U S A. 2006. 103:3840–3845.

15. Goldmacher VS, Bartle LM, Skaletskaya A, Dionne CA, Kedersha NL, Vater CA, et al. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc Natl Acad Sci U S A. 1999. 96:12536–12541.

16. Skaletskaya A, Bartle LM, Chittenden T, McCormick AL, Mocarski ES, Goldmacher VS. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc Natl Acad Sci U S A. 2001. 98:7829–7834.

17. Marshall EE, Bierle CJ, Brune W, Geballe AP. Essential role for either TRS1 or IRS1 in human cytomegalovirus replication. J Virol. 2009. 83:4112–4120.

18. Child SJ, Hakki M, De Niro KL, Geballe AP. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J Virol. 2004. 78:197–205.

19. Colletti KS, Smallenburg KE, Xu Y, Pari GS. Human cytomegalovirus UL84 interacts with an RNA stem-loop sequence found within the RNA/DNA hybrid region of oriLyt. J Virol. 2007. 81:7077–7085.

20. Zhu Y, Huang L, Anders DG. Human cytomegalovirus oriLyt sequence requirements. J Virol. 1998. 72:4989–4996.

21. DePamphilis ML. Transcriptional elements as components of eukaryotic origins of DNA replication. Cell. 1988. 52:635–638.

22. Kagele D, Gao Y, Smallenburg K, Pari GS. Interaction of HCMV UL84 with C/EBPalpha transcription factor binding sites within oriLyt is essential for lytic DNA replication. Virology. 2009. 392:16–23.

23. Kagele D, Rossetto CC, Tarrant MT, Pari GS. Analysis of the interactions of viral and cellular factors with human cytomegalovirus lytic origin of replication, oriLyt. Virology. 2012. 424:106–114.

24. Deng Z, Lezina L, Chen CJ, Shtivelband S, So W, Lieberman PM. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol Cell. 2002. 9:493–503.

25. Wang Y, Li H, Tang Q, Maul GG, Yuan Y. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: involvement of host cellular factors. J Virol. 2008. 82:2867–2882.

26. Xu Y, Cei SA, Rodriguez Huete A, Colletti KS, Pari GS. Human cytomegalovirus DNA replication requires transcriptional activation via an IE2- and UL84-responsive bidirectional promoter element within oriLyt. J Virol. 2004. 78:11664–11677.

27. Huang L, Zhu Y, Anders DG. The variable 3' ends of a human cytomegalovirus oriLyt transcript (SRT) overlap an essential, conserved replicator element. J Virol. 1996. 70:5272–5281.

28. Park MY, Kim YE, Seo MR, Lee JR, Lee CH, Ahn JH. Interactions among four proteins encoded by the human cytomegalovirus UL112-113 region regulate their intranuclear targeting and the recruitment of UL44 to prereplication foci. J Virol. 2006. 80:2718–2727.

29. Staprans SI, Rabert DK, Spector DH. Identification of sequence requirements and trans-acting functions necessary for regulated expression of a human cytomegalovirus early gene. J Virol. 1988. 62:3463–3473.

30. Wright DA, Staprans SI, Spector DH. Four phosphoproteins with common amino termini are encoded by human cytomegalovirus AD169. J Virol. 1988. 62:331–340.

31. Wright DA, Spector DH. Posttranscriptional regulation of a class of human cytomegalovirus phosphoproteins encoded by an early transcription unit. J Virol. 1989. 63:3117–3127.

32. Yamamoto T, Suzuki S, Radsak K, Hirai K. The UL112/113 gene products of human cytomegalovirus which colocalize with viral DNA in infected cell nuclei are related to efficient viral DNA replication. Virus Res. 1998. 56:107–114.

33. Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, et al. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A. 2003. 100:14223–14228.

34. Yu D, Silva MC, Shenk T. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A. 2003. 100:12396–12401.

35. Iwayama S, Yamamoto T, Furuya T, Kobayashi R, Ikuta K, Hirai K. Intracellular localization and DNA-binding activity of a class of viral early phosphoproteins in human fibroblasts infected with human cytomegalovirus (Towne strain). J Gen Virol. 1994. 75:3309–3318.

36. Kerry JA, Priddy MA, Jervey TY, Kohler CP, Staley TL, Vanson CD, et al. Multiple regulatory events influence human cytomegalovirus DNA polymerase (UL54) expression during viral infection. J Virol. 1996. 70:373–382.

37. Li J, Yamamoto T, Ohtsubo K, Shirakata M, Hirai K. Major product pp43 of human cytomegalovirus U(L)112-113 gene is a transcriptional coactivator with two functionally distinct domains. Virology. 1999. 260:89–97.

38. Wells R, Stensland L, Vieira J. The human cytomegalovirus UL112-113 locus can activate the full Kaposi's sarcoma-associated herpesvirus lytic replication cycle. J Virol. 2009. 83:4695–4699.

39. Kim YE, Ahn JH. Role of the specific interaction of UL112-113 p84 with UL44 DNA polymerase processivity factor in promoting DNA replication of human cytomegalovirus. J Virol. 2010. 84:8409–8421.

40. Ahn JH, Jang WJ, Hayward GS. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J Virol. 1999. 73:10458–10471.

41. Penfold ME, Mocarski ES. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology. 1997. 239:46–61.

42. Ertl PF, Powell KL. Physical and functional interaction of human cytomegalovirus DNA polymerase and its accessory protein (ICP36) expressed in insect cells. J Virol. 1992. 66:4126–4133.

43. Weiland KL, Oien NL, Homa F, Wathen MW. Functional analysis of human cytomegalovirus polymerase accessory protein. Virus Res. 1994. 34:191–206.

44. Zuccola HJ, Filman DJ, Coen DM, Hogle JM. The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol Cell. 2000. 5:267–278.

45. Novotny J, Rigoutsos I, Coleman D, Shenk T. In silico structural and functional analysis of the human cytomegalovirus (HHV5) genome. J Mol Biol. 2001. 310:1151–1166.

46. Komazin-Meredith G, Petrella RJ, Santos WL, Filman DJ, Hogle JM, Verdine GL, et al. The human cytomegalovirus UL44 C clamp wraps around DNA. Structure. 2008. 16:1214–1225.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download