Abstract
Cryptococcus gattii causes life-threatening yeast infection in the pulmonary and central nervous systems of humans and animals, and traditionally has been considered to restrict into the tropical and subtropical areas. Despite rare incidence of cryptococcosis caused by C. gattii in Korea, three strains of C. gattii isolated from cryptococcosis patients between 1993 and 2010 were identified. To determine the genetic diversity, 3 strains of C. gattii were typed using PCR fingerprinting with primer M13 and the restriction fragment length polymorphism (RFLP) of orotidine monophosphosphate pyrophosphorylase (URA5) gene. All isolates were identified as serotype B and MATα mating type. The molecular types of each strain, on the other hand, turned out to be distinct belonging to VGI, VGII or III types, respectively. Although the travel histories of the patients were not available, clinical C. gattii strains isolated in Korea may represent the diverse molecular types existing worldwide.
Cryptococcosis is a life-threatening systemic fungal infection affecting healthy and immunocompromised hosts. It is caused by two species of the genus Cryptococcus: C. neoformans and C. gattii (1). Although both species cause either pulmonary or central nervous system infections, they differ in their ecology, epidemiology and clinical manifestations of disease (2, 3). C. neoformans has been isolated worldwide, typically causing disease in immunocompromised hosts, especially AIDS patients. Currently, there are two variants of C. neoformans are recognized, C. neoformans var. grubii (serotype A), which is found worldwide, and C. neoformans var. neoformans (serotype D), which occurs mainly in Europe and South America (4, 5). C. gattii, previously classified as C. neoformans var. gattii (serotypes B and C), was considered to be restricted to tropical and subtropical climates zones, and has been proposed to have a specific ecological association with a number of Eucalyptus species. Also this fungus has a predilection for infecting immunocompetent hosts (6, 7). The relatively uncommon fungal pathogen C. gattii recently emerged as a significant cause of cryptococcal disease in human and animals from temperate regions in the Canada, United States, Europe and Asia (8~10).
The most recent classification of Cryptococcus species was established by molecular typing using PCR fingerprinting, random amplification of polymorphic DNA (RAPD), multilocus sequence typing (MLST) and amplified fragment length polymorphism (AFLP). Based on genetic differences, four distinct molecular types of C. neoformans (VNI to VNIV) and C. gattii (VGI to VGIV) have been recognized by various molecular typing methods (11, 12). These major genotypes differ in their epidemiology, virulence and geographic distribution. The molecular types VGI and VGII are prevalent worldwide and cause the majority of cases in healthy hosts. VGIII, which contains both serotypes B and C, has been isolated from America, Australasia, and Southern Asia, and VGIV, frequently associated with serotype C, is rare globally. VGIII and VGIV appear to more commonly infect immunocompromised patients, including those with AIDS similar to C. neoformans (11, 13, 14). Although VGII is the genotype associated with outbreak in the British Columbia and Western North America, the most common genotype (in Australia and elsewhere) is VGI (9). However, until now Korea has not been considered as C. gattii-endemic area.
C. gattii was isolated from many specimens from not only tropical and subtropical area, but also temperate region. Based on those observations, we thought that ecological and environmental studies on C. gattii in Korea needed to determine reservoirs and to improve understanding of C. gattii epidemiology. In the present study, we investigated the characterization of C. gattii strains isolated from cryptococcosis patients who had been hospitalized between 1993 and 2010 in Korea.
We examined 125 isolates of C. neoformans isolated from 125 patients admitted to 4 province hospitals over 17 years period. Each isolate was purified by picking up the single colony on Saubouraud dextrose agar plate. The purified colonies were tested for their ability to grow and to grow and produce polysaccharide capsule, melanin, and urease at 37℃. The biovariety and serotype of each strain was determined using the canavanine-glycine-bromothymol blue agar test (15) and the Iatron Cryptocheck kit (Iatron Laboratory, Tokyo, Japan), respectively. For comparison, the following reference strains of the known major molecular types of the C. gattii species were included: KACC45444 (serotype B, VGI), KACC45446 (serotype B, VGII), KACC45445 (serotype B, VGIII), KACC45447 (serotype C, VGIV).
DNA was extracted using a Qiagen DNeasy plant mini kit according to the standard protocol. The molecular typing approaches included M13-based PCR fingerprinting and URA5-restriction fragment length polymorphism (RFLP) analysis (16). PCR fingerprinting using the minisatellite-specific core sequence of the wild-type phage M13 (5'-GAGGGTGGCGGTTCT-3') was used as a single primer in the PCR. URA5-RFLP analysis was performed using the primers (5'-ATGTCCTCCCAAGCCCTCGACTCCG-3') and (5'-TTAAGACCTCTGAACACCGTACTC-3'), and RFLP profiles were generated after digestion of the PCR product of the URA5 gene sequence with Sau96l and Hhal. For mating type of C. gattii, we used the mating pheromone-specific primers as described previously (12).
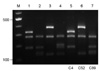
All three isolates were C. gattii serotype B and mating type MATα. Two patients had underlying conditions, diabetes mellitus and cancer, respectively, and the other had no recognizable predisposing factors for cryptococcosis (Table 1). The PCR fingerprinting with the minisatellite-specific primer M13 was able to generate individual strain-specific DNA polymorphism, allowing the easy differentiation among the Cryptococcus species complex. PCR fingerprinting profiles revealed different patterns for C. gattii isolates when compared with reference strains (Fig. 1). Also the URA5 RFLP confirmed each strain to be VGI (C4), VGIII (C52) and VGII (C89) molecular type, respectively (Fig. 2).
Until 1999, most human C. gattii infections were reported from Australia and other tropical and subtropical regions, such as Africa, Southeast Asia, and South America (4, 5). After that C. gattii has acquired the ability to colonize new biogeoclimatic regions and is responsible for a recent outbreak of infection among humans and animals in the temperate climate of Vancouver Island, British Columbia (BC), Canada (9). Even though the reasons are not yet fully understood, its emergence in a temperate climate suggests that the pathogen might have adapted to new climatic niche, or that climatic warming might have created an environment of minimum threshold conditions for C. gattii spore survival. Moreover, the use of immunosuppressive drugs on certain patients, which resulted in increased opportunistic infection of Cryptococcus in immunocompromised hosts may have activated the research of this organism. The data on the global epidemiology and molecular typing of the C. neoformans/C. gattii are well accomplished but limited information on the molecular epidemiology of cryptococcal infections is available from Far East Asia (12, 17). Globally cryptococcosis caused by C. neoformans (serotype A, VNI type) is most common in AIDS and immunocompromised hosts. In 2002, Hwang (18) reported a case of isolation of C. gattii in Korea for the first time, and the major cause of cryptococcosis in Korea is C. neoformans serotype A and the prevalently isolated molecular type is VNI, which shows genetically homogeneous population existing worldwide (12).
In China, located in the Far East Asia, nine C. gattii strains were isolated from 1980 to 2006. All of them were found out to be serotype B, mating type α and VGI type which are commonly isolated in South East Asia. Of the 9 C. gattii strains, 2 were isolated from AIDS patients and 7 were isolated from patients with no apparent risk factors (17). In the present study, 3 molecular types, VGI, VGIII and VGII were identified from each patient with no underlying disease (C4 strain), hepatoma (C52) and diabetes mellitus (C89), respectively. In Japan, Okamoto (3) reported a case of cerebral cryptococcoma caused by C. gattii VGIIa, which is identical to the Vancouver Island outbreak strain, in a patient who had no recent travel history to known disease-endemic areas. Compared to C. neoformans, less information is known about the epidemiology and ecology of C. gattii, especially molecular types VGIII and VGIV (14). Accumulating evidence in recent years indicates that C. gattii is also one of the causes of human cryptococcosis in temperate area including Korea. Although the total number of the indentified C. gattii strains is small, the presence of 3 molecular types recovered in Korea indicates wide geographic distribution of cryptococcosis in Far East Asia.
Considering growth rate of number of cancer patients in Korea, the significance of opportunistic pathogens is also increased. Further characterization and identification of Cryptococcus species should be done to control the infection rate of cryptococcosis in immunocompromised patients and obtain a better understanding of the epidemiology of cryptococcosis in Korea.
Figures and Tables
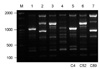 | Figure 1PCR-fingerprint patterns obtained with the primer M13 of Cryptococcus gattii strains from Korea and the reference strains of molecular type. Lanes: 1-4: C. gattii reference strains VGI, VGII, VGIII, and VGIV; lanes: 5-7: Clinical C. gattii strains C4 (VGI), C52 (VGIII), and C89 (VGII). M: molecular marker (100 bp ladder) |
 | Figure 2
URA 5-RFLP profiles obtained after double digestion with the restriction enzymes Hhal and Sau96l of clinical C. gattii strains from Korea and the reference strains of molecular type. Lanes: 1-4: C. gattii reference strains VGI, VGII, VGIII, and VGIV; lanes: 5-7: Clinical C. gattii strains C4 (VGI), C52 (VGIII), and C89 (VGII). M: molecular marker (100 bp ladder). |
References
1. Kwon-Chung KJ, Varma A. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 2006. 6:574–587.

2. Bennett JE, Kwon-Chung KJ, Howard DH. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am J Epidemiol. 1977. 105:582–586.
3. Okamoto K, Hatakeyama S, Itoyama S, Nukui Y, Yoshino Y, Kitazawa T, et al. Cryptococcus gattii genotype VGIIa infection in Man, Japan, 2007. Emerg Infect Dis. 2010. 16:1155–1157.
4. Kwon-Chung KJ, Bennett JE. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am J Epidemiol. 1984. 120:123–130.

5. Franzot SP, Salkin IF, Casadevall A. Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A isolates. J Clin Microbiol. 1999. 37:838–840.

6. Casadevall A, Perfect JR. Cryptococcus neoformans. 1998. Washington: ASM.
7. Ellis DH. Cryptococcus neoformans var. gattii in Australia. J Clin Microbiol. 1987. 25:430–431.
8. Dixit A, Carroll SF, Qureshi ST. Cryptococcus gattii: an emerging cause of fungal disease in North America. Interdiscip Perspect Infect Dis. 2009. 2009:840452.
9. Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A. 2004. 101:17258–17263.

10. Datta K, Bartlett KH, Marr KA. Cryptococcus gattii: emergence in western North America: exploitation of a novel ecological niche. Interdiscip Perspect Infect Dis. 2009. 2009:176532.
11. Meyer W, Marszewska K, Amirmostofian M, Igreja RP, Hardtke C, Methllng K, et al. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA-a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis. 1999. 20:1790–1799.

12. Choi YH, Ngamskulrungroj P, Varma A, Sionov E, Hwang SM, Carriconde F, et al. Prevalence of the VNIc genotype of Cryptococcus neoformans in non-HIV-associated cryptococcosis in the Republic of Korea. FEMS Yeast Res. 2010. 10:769–778.

13. Springer DJ, Chaturvedi V. Projecting global occurrence of Cryptococcus gattii. Emerg Infect Dis. 2010. 16:14–20.
14. Byrnes EJ 3rd, Li W, Ren P, Lewit Y, Voelz K, Fraser JA, et al. A diverse population of Cryptococcus gattii molecular type VGIII in southern California HIV/AIDS patients. PLoS Pathog. 2011. 7:e1002205.
15. Kwon-Chung KJ, Polacheck I, Bennett JE. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C). J Clin Microbiol. 1982. 15:535–537.

16. Meyer W, Castañeda A, Jackson S, Huynh M, Castañeda E. Molecular typing of Ibero American Cryptococcus neoformans isolates. Emerg Infect Dis. 2003. 9:189–195.
17. Chen J, Varma A, Diaz MR, Litvintseva AP, Wollenberg KK, Kwon-Chung KJ. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg Infect Dis. 2008. 14:755–762.

18. Hwang SM. Serotyping of Cryptococcus neoformans strains isolated in Korea. J Microbiol. 2002. 40:166–169.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download