Abstract
Flaviviruses have been important human pathogens after emerging and resurging flavivirus diseases over the past decades. Although effective therapeutic agents are not yet commercially available for use in humans, significant progress has been made toward developing effective therapeutics and treatments. Several studies have shown that antibodies against the flaviviral E and NS1 proteins play a central role in prophylaxis and/or treatment of flavivirus infection through passive immunization. In addition, many anti-flavivirals, including interferons, oligonucleotide-based platforms, and small compounds, have been developed and evaluated for their antiviral effects. This review provides an overview of various approaches to the development of anti-flaviviral candidates and new insights that could improve our strategies for designing effective therapeutics against flaviviruses.
Flavivirus infection has resurged in recent decades and has caused hundreds of thousands of deaths annually worldwide (1~11). The genus Flavivirus of the family Flaviviridae is composed of more than 70 viruses that are transmitted by mosquitoes, ticks, or zoonotic agents with unidentified vectors (8). Approximately 40 viruses in the genus are associated with human diseases. Among these flaviviruses, dengue virus (DENV), West Nile virus (WNV), Japanese encephalitis virus (JEV), yellow fever virus (YFV), St. Louis encephalitis virus (SLEV), and tick-borne encephalitis virus (TBEV) are significant human pathogens globally, which induce severe encephalitic or hemorrhagic diseases that cause extensive morbidity and mortality (6, 8, 12~14).
Flaviviruses are single-stranded, positive-sense, enveloped RNA viruses with an approximately 11-kilobase genome. The genome is translated as a single polyprotein, which is cleaved by viral and cellular proteases to generate three structural proteins [capsid (C), pre-membrane (prM), and envelope (E)] and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (15). Among them, the E protein functions in binding of the virus to a cell surface receptor, membrane fusion, and viral assembly with the assistance of prM (16). Flavivirus NS1 is a glycoprotein that is absent in the virion and secreted at high levels (up to 50 µg/ml) in the serum (17~24). Secreted NS1 associates with the cell surface membrane through interactions with sulfated glycosaminoglycans (25). Additionally, secreted and/or cell-associated NS1 is implicated in the pathogenesis and immune regulation of flavivirus infection in that it binds complement-regulatory factors (26~30). Two other NS proteins, NS3 and NS5, have been characterized as the viral protease/helicase and RNA-dependent RNA polymerase, respectively, which form a viral replication complex with other NS proteins (31, 32).
Although advances in anti-flaviviral drug discovery have progressed significantly, no therapeutic agent for flavivirus infection is currently approved for human use. Recent studies suggest that monoclonal antibody (mAb)-based therapy could be a promising alternative strategy (33~36). Several groups have generated protective mAbs against the E and NS1 proteins of flaviviruses (33, 34, 37~44). The structural E protein is used as the major antigenic target to raise neutralizing antibodies (33, 35, 36, 45). However, it is difficult to generate neutralizing antibody for variant viruses due to the high mutation rate of the flavivirus RNA-dependent RNA polymerase (46). Sub-neutralizing concentrations of anti-E antibody also have theoretical potential to cause antibody-dependent enhancement (ADE) of flavivirus infection (47~50). Another major antigen, the flavivirus NS1 glycoprotein, induces the production of non-neutralizing protective antibodies (34, 51~53). Although the protective mechanisms and binding regions of a few anti-NS1 antibodies have been demonstrated (24, 34, 54~56), most still remain to be characterized. On the other hand, several antiviral agents have been tested to overcome the drawback for anti-flavivirus antibodies (57~63). Some of these agents inhibit flavivirus infection and reduce the significant mortality and morbidity associated with the infections (62, 63).
This review provides an overview of the major antiflavivirus antibodies and other anti-flaviviral agents with particular attention to: [1] protective and/or therapeutic antibodies against flavivirus infections, [2] the inhibitory mechanisms of protective and/or therapeutic antibodies, and [3] other candidates for anti-flavivirus therapeutics.
Flavivirus infection induces the humoral immune response in the host and elicits the production of anti-flavivirus antibodies that could limit viral spread and burden (64~69). As expected, many studies demonstrate that passive administration of polyclonal or monoclonal antibodies against flavivirus proteins protects mice from lethal flavivirus infection (33, 34, 37~44), and B-cell-deficient mice are more vulnerable to infection (66). These findings indicate that antibodies are one of the major components involved in protection against flavivirus infection.
A flavivirus virion consists of three structural proteins (C, prM/M, and E), the viral genome, and a lipid envelope derived from the endoplasmic reticulum (ER) (70, 71). The viral particle induces the production of neutralizing antibodies (72~74). Although some neutralizing antibodies recognize the prM/M protein, the majority of neutralizing antibodies are raised against the E protein (75~77). The E protein has three structural domains and plays an important role in viral attachment, entry, assembly, and cell tropism (Fig. 1) (16). Domain I (DI) and Domain II (DII) of the E protein are involved in pH-dependent fusion of the virus and host cell membranes (78). Domain III (DIII) has an immunoglobulin-like fold and is located on the opposite end of DI, which is suggested to contain cellular receptor binding sites (43, 79, 80). Although neutralizing mAbs that cross-react with flaviviruses primarily recognize DII, most neutralizing antibodies bind to epitopes in the DIII region (Fig. 1) (43, 44, 81~83). In addition, X-ray crystallography and neutralization escape mutant analysis indicate that type-specific neutralizing antibodies against individual flaviviruses mainly map to amino acid residues in DIII of the E protein (Fig. 1) (84~87).
Interestingly, the anti-E neutralizing mAbs at sub-neutralizing concentrations have the potential to result in ADE of flavivirus infections, thereby complicating antibody therapy (47~50). Although ADE by anti-E neutralizing mAbs has not been well characterized in flavivirus infection in vivo, in vitro studies suggest that ADE is primarily associated with Fc-γ- or complement receptors (48, 88~90). To avoid the potential ADE, recent studies engineered anti-flavivirus antibodies with mutations in the Fc region, which prevented ADE in vitro and in vivo (88). Therefore, these findings suggest that ADE of flavivirus infection is a serious consideration in the design of novel strategies to develop a safe and effective therapeutic agent based on anti-E antibodies.
During the course of natural infection, flaviviruses secrete nonstructural protein NS1, which is present at high concentrations (e.g., 1~50 µg/ml) in patient serum and associates with cell surface membranes (20~23, 25). Although most neutralizing antibodies are raised against virion-associated proteins, E and prM/M, NS1, which is absent from the virion, induces the production of non-neutralizing protective antibodies. Many studies have demonstrated the immunogenicity and protective efficiency of recombinant NS1 generated through DNA vaccines, recombinant viruses, and bacterial expression (38, 39, 51, 52, 91). The passive administration of anti-NS1 antibody protects mice against lethal flavivirus challenge, depending on the dosage and time of administration (34). These results suggest that anti-NS1 antibodies could serve as therapeutic antibodies that do not induce ADE. However, a recent study revealed that anti-DENV-2 NS1 mAbs (e.g., 1G5.3) cross-reacts with the DENV-2 E protein, inducing weak neutralizing activity and ADE in mice. These findings imply a potential risk of anti-NS1 antibody-based therapeutics (92).
Because mapping of protective mAbs could provide useful information for the design of effective therapeutics, several studies have been performed to determine the binding regions for anti-NS1 mAbs by using overlapping peptides, bacterially expressed fragments of NS1, yeast surface display expression, and mAb competition binding assays (24, 34, 54~56). Despite the intense interest, few protective mAbs against NS1 have been mapped to specific amino acids (53, 54, 91) and the three-dimensional structure of the NS1 protein has not yet been identified. Recently, important determinants for a cross-protective mAb against JEV and WNV, 16NS1, were identified by overlapping peptide mapping analysis combined with a yeast surface display system and site-specific mutagenesis (53). However, structural studies based on X-ray crystallography are required to further characterize the functions of flavivirus NS1 and protective anti-NS1 antibodies.
Protective antibodies prevent viral infection and reduce the viral burden in host cells through direct and indirect effects (33~35, 45). Neutralization is one of the direct functions of antibodies that does not require any other immune system components and is independent of the Fc portion of the antibody. Neutralizing antibodies block many steps in the viral entry pathway including virion attachment to the host cell, entry into the host cell, and uncoating in the endosome to release viral RNA into cytoplasm (Fig. 2) (93~96). For example, E53 and E60 anti-WNV E mAbs block viral attachment and entry at neutralizing concentrations (Fig. 2A) (96). The E16 anti-WNV E mAb inhibits fusion of WNV with the endosomal membrane and blocks uncoating, which leads the virus particle to the lysosome for destruction (Fig. 2B) (96). Although some neutralizing antibodies are well characterized, our understanding of neutralization is still limited. For example, an anti-DNEV2 mAb, 3H5-1, inhibits viral attachment to Vero cells, but the 3H5-1 mAb blocks DENV-2 fusion to the plasma membrane of LLC-MK2 cells (97, 98). In addition, while attachment factors (e.g., DC-SIGN, DC-SIGNR, heparin sulfate) that facilitate flavivirus binding in viral entry were identified (99~101), a neutralizing antibody that inhibits the E-attachment factor complex has not yet been characterized.
Non-neutralizing antibodies exert a protective effect through indirect functions that require the Fc portion of the antibody and components of the innate or adaptive immune system (102). Despite the absence of detectable neutralizing activity of the anti-NS1 antibody, many studies have reported its protective activity (34, 37, 39, 40, 42, 53, 91). Although the detailed mechanisms of this protection are incompletely understood, antiviral functions through an Fc-dependent pathway and complement-mediated cytolysis (CMC) of infected cells have been proposed as the basis for mAbs protection against NS1 (Fig. 3) (34, 42, 103). For example, anti-NS1 antibody bound to YFV-infected cells induces CMC of virus-infected cells (Fig. 3A) (103). Recent passive antibody transfer studies showed that anti-WNV NS1 mAbs (10NS1, 16NS1, and 17NS1) trigger protective activity through a C1q-independence and Fc-γ receptor I-and/or IV-mediated phagocytosis (Figs. 3C and D) (34, 37). However, specific mechanisms of many non-neutralizing antiviral antibodies against flavivirus NS1 remain to be identified. For example, an anti-WNV NS1 mAb, 14NS1, confers a strong protective effect in mice infected with lethal WNV that are deficient in C1q and Fc-γ receptor I and III (34), but the detailed mechanism underlying this effect remains uncharacterized. Although more studies on the detailed mechanisms of these anti-NS1 mAbs are needed, the facts that the cell surface-associated NS1 of WNV modulates complement activation by binding complement regulatory protein and the secreted NS1 of DENV increases viral propagation indicate mAbs to flavivirus NS1 may directly block the immunomodulatory and virologic functions of NS1 (26, 104).
Beyond direct neutralization of anti-flavivirus E mAbs, recent observations suggest that the protective activity of some neutralizing mAbs is partially dependent on the Fc portion of the mAbs and is associated with Fc receptors and the complement cascade (Fig. 3B) (48, 82, 105). For example, the protective efficiency of an anti-WNV E mAb is reduced in mice with blocked Fc-γ receptors I, III, and IV (82). The neutralization potential of hu-E16 anti-WNV E is augmented by C1q, which is dependent on the isotype of antibodies that bind C1q avidly (human IgG1 and IgG3) (48). Taken together, these recent findings suggest that a better comprehension of the role on protective antibodies in vivo and in vitro is crucial for the development of optimal therapeutic antibodies.
Although antibody-based therapy provides a promising strategy to inhibit flavivirus infection, one possible limitation of this therapeutic antibody is the emergence of escape mutants that could decrease the inhibitory activity (46). Several antiviral agents have been tested and characterized to determine their inhibitory activity against viral replication in host cells (57~63). However, it is more difficult to develop specific antiviral agents without toxicity to cells because viruses, unlike bacteria, are obligate intracellular parasites that are dependent on the host's biosynthetic machinery. In this section, several anti-flaviviral agents will be discussed.
Interferons are produced through the innate immune response to viral infection (106). During the viral life cycle, double-stranded viral RNA can primarily induce type I interferons such as interferon-α and interferon-β in the infected cell. Type I interferons inhibit viral replication by activating the JAK-STAT pathway, which induces the expression of antiviral genes. The interferon-dependent innate immune response is crucial for inhibiting flavivirus infection (107~111). For example, interferon-α/β receptor-deficient mice are more susceptible to WNV infection with 100% mortality and high viral loads in nearly all tissues (111). Type I interferon-pretreatment of cells also significantly inhibits flaviviruses (107~109, 111). However, the antiviral activity of interferon is significantly reduced after viral replication because flavivirus nonstructural proteins interfere with interferon signaling pathway (110, 112, 113). Nonetheless, several lines of evidence indicate that interferon has strong potential for use as a therapeutic. For example, treatment with interferon-α yields substantial improvement in complications in SLEV and WNV encephalitis cases (114, 115).
Nucleic acid-based antiviral agents involve the use of oligonucleotides to suppress viral gene expression, including antisense-, ribozyme- and RNA interference (RNAi)-based approaches (116, 117). These strategies selectively inhibit viral replication by targeting the expression of key viral proteins through degradation of sequence-specific single-stranded RNA (118). RNAi is an evolutionarily conserved cellular mechanism that is initiated by double-stranded RNA (dsRNA) or micro RNA, which specifically blocks gene expression (119, 120). In the past decade, RNAi has been widely used to inhibit flavivirus infection in cells (57, 59, 121, 122). For example, small interfering RNA (siRNA) inhibits JEV replication (121). In addition, pretreatment of siRNA prior to viral replication significantly reduces WNV infection (59, 122). Recent studies show that administration of siRNA improves survival against lethal flavivirus infection in mice (62, 63). Although these results suggest that antiviral RNAi therapy is very promising, the emergence of escape mutations in the targeted sequences may limit their efficiency. However, the development of a new delivery method could increase their therapeutic potential and clinical applications because commonly used methods such as electroporation, lipid-based transfection reagents, and nanoparticles are less effective and not cell specific.
Viral replication is a well-organized process that is essential for effectively producing progeny viruses. Therefore, the steps of viral replication could be attractive targets for antiviral agents that inhibit viral genome replication and enzymes whose activity is crucial for viral protein processing (123). Two nonstructural proteins with enzymatic functions, NS3 (protease and helicase) and NS5 (RNA-dependent RNA polymerase), are considered as major targets for antiviral inhibitors, and a small-molecule library has been screened to identify compounds that block viral enzymes critical for replication (124~127). Borowski et al. demonstrated that an imidazo[4,5-d]pyridazine nucleoside analogue, 1-(2'-Omethyl-β-D-ribofuranosyl)imidazo[4,5-d]pyridazine-4,7(5H, 6H)-dione, inhibits the helicase activity of WNV NS3 (IC50 = 30 µM) (124). In addition, this nucleotide analogue shows similar inhibition against WNV replication in cell culture (125). Johnston et al. screened a compound library to identify inhibitors of WNV NS2B-NS3 protease using a miniature NS2B-NS3 assay and discovered a common amino-1H-pyrazol-3-yl scaffold as an inhibitor of WNV protease (126). Migliaccio et al. reported that nucleoside analogs, 2'-C-methyl-substituted ribonucleosides, inhibit flaviviruses such as WNV, DENV, and YFV by termination of RNA synthesis (127).
Several groups have recently used a high-throughput screening (HTS) assay combined with a flavivirus replicon and/or virus-like particle (VLP) system to discover novel flavivirus inhibitors (128, 129). Qing et al. developed an HTS assay using DENV-1 VLP to screen anti-DENV-1 inhibitors (128). Noueiry et al. employed a cell-based WNV subgenomic replicon to test over 80,000 compounds to determine their capacity to inhibit WNV replication and identified lead compound classes with strong antiviral activity against WNV, including 5H-cyclopenta [b] pyridine, tert-sulfonamide, thienylpyrimidine, and secondary sulfonamides (129). Approaches based on these HTS assays could facilitate the development of anti-flavivirus drugs that target viral entry, translation, and replication.
During the past decades, the resurgence and spread of the most important mosquito-borne flaviviruses (e.g., DENV, WNV, and JEV) to new environments have increased the importance of developing anti-flavivirus agents. Anti-flaviviral candidates with different targets and inhibitory mechanisms have been developed. mAbs against the E and NS1 protein have provided significant protection through neutralization and effector-mediated clearance of virions and infected cells. In addition, other anti-flaviviral agents including type I interferons, oligonucleotide-based platforms, and small compounds targeting flaviviral replication have been developed to inhibit flaviviral infection in vitro and/or in vivo. Although these flavivirus antivirals demonstrate significant prophylactic and/or therapeutic effects against flaviviral infection, combination treatments are required to minimize the risk of escape mutant emergence and to increase the antiviral efficiency through different targets. Furthermore, expanding flavivirus epidemics and the co-circulation of multiple flaviviruses in the same endemic area (7~9) has heightened the necessity for developing broad-spectrum therapeutics against flavivirus infection.
Figures and Tables
Figure 1
Schematic of flavivirus E and its antibody. The structure of the dimer of flavivirus E protein is schematically represented. "I", "II", and "III" represent Domain I, Domain II, and Domain III, respectively. Domain I and II participate in the pH-dependent fusion of virus-host cells, and Domain III has been suggested as a host receptor binding site. Although most neutralizing monoclonal antibodies (mAbs) recognize Domain III, broadly cross-neutralizing flavivirus mAbs primarily react with Domain II.
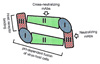
Figure 2
Protection model of neutralizing antibody. (A) Blockage of virion attachment and entry. Neutralizing antibodies that are directed against the E protein inhibit virion attachment and entry by blocking receptor engagement or membrane fusion. (B) Inhibition of virion uncoating. Some neutralizing antibodies interfere with fusion of the virion and endosomal membrane, which results in lysosomal destruction of the virion.
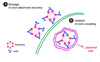
Figure 3
Protection model of non-neutralizing antibody through immune mechanisms. (A) Complement-mediated cytolysis of infected cells. Antibodies bound to a specific antigen on flavivirus-infected cells interact with C1q complement factor, which leads to activation of the classical complement pathway and eventually induces membrane attack complex (MAC)-mediated lysis of virus-infected cells. (B) Antibody-mediated complement lysis of virions. The antiviral effects of some neutralizing antibodies are efficiently enhanced by inducing lysis of the virion through antibody-mediated complement activation, which leads to fragmentation of the viral envelope. (C) and (D) Antibody-dependent clearance of viral particles and infected cells through Fc-γ receptor(s). Antibody binding to viral particles and infected cells recruits immune-effect cells such as macrophages, which interact with the Fc region of the antibody through the Fc-γ receptor expressed by the effect cells. The antibody-bound virions and infected cells are then phagocytosed by the immune cells.
References
1. Lanciotti RS, Roehrig JT, Deubel V, Smith J, Parker M, Steele K, et al. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science. 1999. 286:2333–2337.

2. Mackenzie JS, Barrett AD, Deubel V. The Japanese encephalitis serological group of flaviviruses: a brief introduction to the group. Curr Top Microbiol Immunol. 2002. 267:1–10.

3. Mackenzie JS, Johansen CA, Ritchie SA, van den Hurk AF, Hall RA. Japanese encephalitis as an emerging virus: the emergence and spread of Japanese encephalitis virus in Australasia. Curr Top Microbiol Immunol. 2002. 267:49–73.

4. Lanciotti RS, Ebel GD, Deubel V, Kerst AJ, Murri S, Meyer R, et al. Complete genome sequences and phylogenetic analysis of West Nile virus strains isolated from the United States, Europe, and the Middle East. Virology. 2002. 298:96–105.

5. Jia XY, Briese T, Jordan I, Rambaut A, Chi HC, Mackenzie JS, et al. Genetic analysis of West Nile New York 1999 encephalitis virus. Lancet. 1999. 354:1971–1972.

7. Douglas KO, Kilpatrick AM, Levett PN, Lavoie MC. A quantitative risk assessment of West Nile virus introduction into Barbados. West Indian Med J. 2007. 56:394–397.
8. Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004. 10:S98–S109.

9. Jupp PG. The ecology of West Nile virus in South Africa and the occurrence of outbreaks in humans. Ann N Y Acad Sci. 2001. 951:143–152.

10. Yu Y. Phenotypic and genotypic characteristics of Japanese encephalitis attenuated live vaccine virus SA14-14-2 and their stabilities. Vaccine. 2010. 28:3635–3641.

12. Rodenhuis-Zybert IA, Wilschut J, Smit JM. Partial maturation: an immune-evasion strategy of dengue virus? Trends Microbiol. 2011. 19:248–254.

13. Hubálek Z, Halouzka J. West Nile fever--a reemerging mosquito-borne viral disease in Europe. Emerg Infect Dis. 1999. 5:643–650.

15. Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990. 44:649–688.

16. Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005. 3:13–22.

17. Macdonald J, Tonry J, Hall RA, Williams B, Palacios G, Ashok MS, et al. NS1 protein secretion during the acute phase of West Nile virus infection. J Virol. 2005. 79:13924–13933.

18. Mason PW. Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology. 1989. 169:354–364.

19. Wengler G, Gross HJ. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology. 1978. 89:423–437.

20. Winkler G, Maxwell SE, Ruemmler C, Stollar V. Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology. 1989. 171:302–305.

21. Alcon S, Talarmin A, Debruyne M, Falconar A, Deubel V, Flamand M. Enzyme-linked immunosorbent assay specific to Dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J Clin Microbiol. 2002. 40:376–381.

22. Chung KM, Diamond MS. Defining the levels of secreted non-structural protein NS1 after West Nile virus infection in cell culture and mice. J Med Virol. 2008. 80:547–556.

23. Libraty DH, Young PR, Pickering D, Endy TP, Kalayanarooj S, Green S, et al. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J Infect Dis. 2002. 186:1165–1168.

24. Young PR. Antigenic analysis of dengue virus using monoclonal antibodies. Southeast Asian J Trop Med Public Health. 1990. 21:646–651.
25. Avirutnan P, Zhang L, Punyadee N, Manuyakorn A, Puttikhunt C, Kasinrerk W, et al. Secreted NS1 of dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate E. PLoS Pathog. 2007. 3:e183.

26. Chung KM, Liszewski MK, Nybakken G, Davis AE, Townsend RR, Fremont DH, et al. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc Natl Acad Sci U S A. 2006. 103:19111–19116.

27. Schlesinger JJ. Flavivirus nonstructural protein NS1: complementary surprises. Proc Natl Acad Sci U S A. 2006. 103:18879–18880.

28. Kurosu T, Chaichana P, Yamate M, Anantapreecha S, Ikuta K. Secreted complement regulatory protein clusterin interacts with dengue virus nonstructural protein 1. Biochem Biophys Res Commun. 2007. 362:1051–1056.

29. Avirutnan P, Fuchs A, Hauhart RE, Somnuke P, Youn S, Diamond MS, et al. Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. J Exp Med. 2010. 207:793–806.

30. Avirutnan P, Hauhart RE, Somnuke P, Blom AM, Diamond MS, Atkinson JP. Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation. J Immunol. 2011. 187:424–433.

31. Khromykh AA, Sedlak PL, Guyatt KJ, Hall RA, Westaway EG. Efficient trans-complementation of the flavivirus kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J Virol. 1999. 73:10272–10280.

32. Khromykh AA, Sedlak PL, Westaway EG. cis- and trans-acting elements in flavivirus RNA replication. J Virol. 2000. 74:3253–3263.

33. Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, et al. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat Med. 2005. 11:522–530.

34. Chung KM, Nybakken GE, Thompson BS, Engle MJ, Marri A, Fremont DH, et al. Antibodies against West Nile Virus nonstructural protein NS1 prevent lethal infection through Fc gamma receptor-dependent and -independent mechanisms. J Virol. 2006. 80:1340–1351.

35. Shrestha B, Brien JD, Sukupolvi-Petty S, Austin SK, Edeling MA, Kim T, et al. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathog. 2010. 6:e1000823.

36. Sukupolvi-Petty S, Austin SK, Engle M, Brien JD, Dowd KA, Williams KL, et al. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J Virol. 2010. 84:9227–9239.

37. Chung KM, Thompson BS, Fremont DH, Diamond MS. Antibody recognition of cell surface-associated NS1 triggers Fc-gamma receptor-mediated phagocytosis and clearance of West Nile Virus-infected cells. J Virol. 2007. 81:9551–9555.

38. Desprès P, Dietrich J, Girard M, Bouloy M. Recombinant baculoviruses expressing yellow fever virus E and NS1 proteins elicit protective immunity in mice. J Gen Virol. 1991. 72:2811–2816.

39. Gould EA, Buckley A, Barrett AD, Cammack N. Neutralizing (54K) and non-neutralizing (54K and 48K) monoclonal antibodies against structural and non-structural yellow fever virus proteins confer immunity in mice. J Gen Virol. 1986. 67:591–595.

40. Henchal EA, Henchal LS, Schlesinger JJ. Synergistic interactions of anti-NS1 monoclonal antibodies protect passively immunized mice from lethal challenge with dengue 2 virus. J Gen Virol. 1988. 69:2101–2107.

41. Schlesinger JJ, Brandriss MW, Walsh EE. Protection against 17D yellow fever encephalitis in mice by passive transfer of monoclonal antibodies to the nonstructural glycoprotein gp48 and by active immunization with gp48. J Immunol. 1985. 135:2805–2809.
42. Schlesinger JJ, Foltzer M, Chapman S. The Fc portion of antibody to yellow fever virus NS1 is a determinant of protection against YF encephalitis in mice. Virology. 1993. 192:132–141.

43. Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J Virol. 2001. 75:7769–7773.

44. Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken GE, Schlesinger JJ, et al. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J Virol. 2007. 81:12816–12826.

45. Roehrig JT, Staudinger LA, Hunt AR, Mathews JH, Blair CD. Antibody prophylaxis and therapy for flavivirus encephalitis infections. Ann N Y Acad Sci. 2001. 951:286–297.

46. Khromykh AA, Varnavski AN, Sedlak PL, Westaway EG. Coupling between replication and packaging of flavivirus RNA: evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J Virol. 2001. 75:4633–4640.

47. Cardosa MJ, Gordon S, Hirsch S, Springer TA, Porterfield JS. Interaction of West Nile virus with primary murine macrophages: role of cell activation and receptors for antibody and complement. J Virol. 1986. 57:952–959.

48. Mehlhop E, Ansarah-Sobrinho C, Johnson S, Engle M, Fremont DH, Pierson TC, et al. Complement protein C1q inhibits antibody-dependent enhancement of flavivirus infection in an IgG subclass-specific manner. Cell Host Microbe. 2007. 2:417–426.

49. Peiris JS, Gordon S, Unkeless JC, Porterfield JS. Monoclonal anti-Fc receptor IgG blocks antibody enhancement of viral replication in macrophages. Nature. 1981. 289:189–191.

50. Pierson TC, Xu Q, Nelson S, Oliphant T, Nybakken GE, Fremont DH, et al. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe. 2007. 1:135–145.

51. Lin YL, Chen LK, Liao CL, Yeh CT, Ma SH, Chen JL, et al. DNA immunization with Japanese encephalitis virus nonstructural protein NS1 elicits protective immunity in mice. J Virol. 1998. 72:191–200.

52. Falgout B, Bray M, Schlesinger JJ, Lai CJ. Immunization of mice with recombinant vaccinia virus expressing authentic dengue virus nonstructural protein NS1 protects against lethal dengue virus encephalitis. J Virol. 1990. 64:4356–4363.

53. Lee TH, Song BH, Yun SI, Woo HR, Lee YM, Diamond MS, et al. A cross-protective mAb recognizes a novel epitope within the flavivirus NS1 protein. J Gen Virol. 2012. 93:20–26.

54. Falconar AK, Young PR, Miles MA. Precise location of sequential dengue virus subcomplex and complex B cell epitopes on the nonstructural-1 glycoprotein. Arch Virol. 1994. 137:315–326.

55. Henchal EA, Henchal LS, Thaisomboonsuk BK. Topological mapping of unique epitopes on the dengue-2 virus NS1 protein using monoclonal antibodies. J Gen Virol. 1987. 68:845–851.

56. Putnak JR, Charles PC, Padmanabhan R, Irie K, Hoke CH, Burke DS. Functional and antigenic domains of the dengue-2 virus nonstructural glycoprotein NS-1. Virology. 1988. 163:93–103.

57. Adelman ZN, Sanchez-Vargas I, Travanty EA, Carlson JO, Beaty BJ, Blair CD, et al. RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J Virol. 2002. 76:12925. 12933.

58. Borowski P, Deinert J, Schalinski S, Bretner M, Ginalski K, Kulikowski T, et al. Halogenated benzimidazoles and benzotriazoles as inhibitors of the NTPase/helicase activities of hepatitis C and related viruses. Eur J Biochem. 2003. 270:1645–1653.

59. McCown M, Diamond MS, Pekosz A. The utility of siRNA transcripts produced by RNA polymerase i in down regulating viral gene expression and replication of negative- and positive-strand RNA viruses. Virology. 2003. 313:514–524.

60. Zhang N, Chen HM, Koch V, Schmitz H, Minczuk M, Stepien P, et al. Potent inhibition of NTPase/helicase of the West Nile Virus by ring-expanded ("fat") nucleoside analogues. J Med Chem. 2003. 46:4776–4789.

61. Bai F, Town T, Pradhan D, Cox J, Ashish , Ledizet M, et al. Antiviral peptides targeting the west nile virus envelope protein. J Virol. 2007. 81:2047–2055.

62. Bai F, Wang T, Pal U, Bao F, Gould LH, Fikrig E. Use of RNA interference to prevent lethal murine west nile virus infection. J Infect Dis. 2005. 191:1148–1154.

63. Kumar P, Lee SK, Shankar P, Manjunath N. A single siRNA suppresses fatal encephalitis induced by two different flaviviruses. PLoS Med. 2006. 3:e96.

64. Chambers TJ, Diamond MS. Pathogenesis of flavivirus encephalitis. Adv Virus Res. 2003. 60:273–342.

65. Diamond MS. Evasion of innate and adaptive immunity by flaviviruses. Immunol Cell Biol. 2003. 81:196–206.

66. Diamond MS, Shrestha B, Marri A, Mahan D, Engle M. B cells and antibody play critical roles in the immediate defense of disseminated infection by West Nile encephalitis virus. J Virol. 2003. 77:2578–2586.

67. Diamond MS, Shrestha B, Mehlhop E, Sitati E, Engle M. Innate and adaptive immune responses determine protection against disseminated infection by West Nile encephalitis virus. Viral Immunol. 2003. 16:259–278.

68. Diamond MS, Sitati EM, Friend LD, Higgs S, Shrestha B, Engle M. A critical role for induced IgM in the protection against West Nile virus infection. J Exp Med. 2003. 198:1853–1862.

70. Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002. 108:717–725.

71. Mukhopadhyay S, Kim BS, Chipman PR, Rossmann MG, Kuhn RJ. Structure of West Nile virus. Science. 2003. 302:248.

72. Lim CK, Takasaki T, Kotaki A, Kurane I. Vero cell-derived inactivated West Nile (WN) vaccine induces protective immunity against lethal WN virus infection in mice and shows a facilitated neutralizing antibody response in mice previously immunized with Japanese encephalitis vaccine. Virology. 2008. 374:60–70.

73. Widman DG, Ishikawa T, Fayzulin R, Bourne N, Mason PW. Construction and characterization of a second-generation pseudoinfectious West Nile virus vaccine propagated using a new cultivation system. Vaccine. 2008. 26:2762–2771.

74. Yu L, Robert Putnak J, Pletnev AG, Markoff L. Attenuated West Nile viruses bearing 3'SL and envelope gene substitution mutations. Vaccine. 2008. 26:5981–5988.

75. Colombage G, Hall R, Pavy M, Lobigs M. DNA-based and alphavirus-vectored immunisation with prM and E proteins elicits long-lived and protective immunity against the flavivirus, Murray Valley encephalitis virus. Virology. 1998. 250:151–163.

76. Falconar AK. Identification of an epitope on the dengue virus membrane (M) protein defined by cross-protective monoclonal antibodies: design of an improved epitope sequence based on common determinants present in both envelope (E and M) proteins. Arch Virol. 1999. 144:2313–2330.

77. Pincus S, Mason PW, Konishi E, Fonseca BA, Shope RE, Rice CM, et al. Recombinant vaccinia virus producing the prM and E proteins of yellow fever virus protects mice from lethal yellow fever encephalitis. Virology. 1992. 187:290–297.

78. Allison SL, Schalich J, Stiasny K, Mandl CW, Heinz FX. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J Virol. 2001. 75:4268–4275.

79. Bhardwaj S, Holbrook M, Shope RE, Barrett AD, Watowich SJ. Biophysical characterization and vector-specific antagonist activity of domain III of the tick-borne flavivirus envelope protein. J Virol. 2001. 75:4002–4007.

80. Chu JJ, Rajamanonmani R, Li J, Bhuvanakantham R, Lescar J, Ng ML. Inhibition of West Nile virus entry by using a recombinant domain III from the envelope glycoprotein. J Gen Virol. 2005. 86:405–412.

81. Crill WD, Chang GJ. Localization and characterization of flavivirus envelope glycoprotein cross-reactive epitopes. J Virol. 2004. 78:13975–13986.

82. Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, et al. Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein. J Virol. 2006. 80:12149–12159.

83. Stiasny K, Kiermayr S, Holzmann H, Heinz FX. Cryptic properties of a cluster of dominant flavivirus cross-reactive antigenic sites. J Virol. 2006. 80:9557–9568.

84. Beasley DW, Barrett AD. Identification of neutralizing epitopes within structural domain III of the West Nile virus envelope protein. J Virol. 2002. 76:13097–13100.

85. Chambers TJ, Halevy M, Nestorowicz A, Rice CM, Lustig S. West Nile virus envelope proteins: nucleotide sequence analysis of strains differing in mouse neuroinvasiveness. J Gen Virol. 1998. 79:2375–2380.

86. Hiramatsu K, Tadano M, Men R, Lai CJ. Mutational analysis of a neutralization epitope on the dengue type 2 virus (DEN2) envelope protein: monoclonal antibody resistant DEN2/DEN4 chimeras exhibit reduced mouse neurovirulence. Virology. 1996. 224:437–445.

87. Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005. 437:764–769.

88. Balsitis SJ, Williams KL, Lachica R, Flores D, Kyle JL, Mehlhop E, et al. Lethal antibody enhancement of dengue disease in mice is prevented by Fc modification. PLoS Pathog. 2010. 6:e1000790.

89. Cardosa MJ, Porterfield JS, Gordon S. Complement receptor mediates enhanced flavivirus replication in macrophages. J Exp Med. 1983. 158:258–263.

90. Gollins SW, Porterfield JS. Flavivirus infection enhancement in macrophages: radioactive and biological studies on the effect of antibody on viral fate. J Gen Virol. 1984. 65:1261–1272.

91. Falconar AK. Antibody responses are generated to immunodominant ELK/KLE-type motifs on the nonstructural-1 glycoprotein during live dengue virus infections in mice and humans: implications for diagnosis, pathogenesis, and vaccine design. Clin Vaccine Immunol. 2007. 14:493–504.

92. Falconar AK. Monoclonal antibodies that bind to common epitopes on the dengue virus type 2 nonstructural-1 and envelope glycoproteins display weak neutralizing activity and differentiated responses to virulent strains: implications for pathogenesis and vaccines. Clin Vaccine Immunol. 2008. 15:549–561.

93. Gollins SW, Porterfield JS. A new mechanism for the neutralization of enveloped viruses by antiviral antibody. Nature. 1986. 321:244–246.

94. Roehrig JT, Bolin RA, Kelly RG. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology. 1998. 246:317–328.

95. Stiasny K, Brandler S, Kössl C, Heinz FX. Probing the flavivirus membrane fusion mechanism by using monoclonal antibodies. J Virol. 2007. 81:11526–11531.

96. Thompson BS, Moesker B, Smit JM, Wilschut J, Diamond MS, Fremont DH. A therapeutic antibody against west nile virus neutralizes infection by blocking fusion within endosomes. PLoS Pathog. 2009. 5:e1000453.

97. He RT, Innis BL, Nisalak A, Usawattanakul W, Wang S, Kalayanarooj S, et al. Antibodies that block virus attachment to Vero cells are a major component of the human neutralizing antibody response against dengue virus type 2. J Med Virol. 1995. 45:451–461.

98. Se-Thoe SY, Ling AE, Ng MM. Alteration of virus entry mode: a neutralisation mechanism for Dengue-2 virus. J Med Virol. 2000. 62:364–376.

99. Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ, et al. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997. 3:866–871.

100. Davis CW, Nguyen HY, Hanna SL, Sánchez MD, Doms RW, Pierson TC. West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection. J Virol. 2006. 80:1290–1301.

101. Su CM, Liao CL, Lee YL, Lin YL. Highly sulfated forms of heparin sulfate are involved in japanese encephalitis virus infection. Virology. 2001. 286:206–215.

102. Law M, Hangartner L. Antibodies against viruses: passive and active immunization. Curr Opin Immunol. 2008. 20:486–492.

103. Schlesinger JJ, Brandriss MW, Putnak JR, Walsh EE. Cell surface expression of yellow fever virus non-structural glycoprotein NS1: consequences of interaction with antibody. J Gen Virol. 1990. 71:593–599.

104. Alcon-LePoder S, Drouet MT, Roux P, Frenkiel MP, Arborio M, Durand-Schneider AM, et al. The secreted form of dengue virus nonstructural protein NS1 is endocytosed by hepatocytes and accumulates in late endosomes: implications for viral infectivity. J Virol. 2005. 79:11403–11411.

105. Gould LH, Sui J, Foellmer H, Oliphant T, Wang T, Ledizet M, et al. Protective and therapeutic capacity of human single-chain Fv-Fc fusion proteins against West Nile virus. J Virol. 2005. 79:14606–14613.

106. Samuel CE. Antiviral actions of interferon. Interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology. 1991. 183:1–11.

107. Anderson JF, Rahal JJ. Efficacy of interferon alpha-2b and ribavirin against West Nile virus in vitro. Emerg Infect Dis. 2002. 8:107–108.

108. Crance JM, Scaramozzino N, Jouan A, Garin D. Interferon, ribavirin, 6-azauridine and glycyrrhizin: antiviral compounds active against pathogenic flaviviruses. Antiviral Res. 2003. 58:73–79.

109. Diamond MS, Harris E. Interferon inhibits dengue virus infection by preventing translation of viral RNA through a PKR-independent mechanism. Virology. 2001. 289:297–311.

110. Diamond MS, Roberts TG, Edgil D, Lu B, Ernst J, Harris E. Modulation of Dengue virus infection in human cells by alpha, beta, and gamma interferons. J Virol. 2000. 74:4957–4966.

111. Samuel MA, Diamond MS. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J Virol. 2005. 79:13350–13361.

112. Lin RJ, Liao CL, Lin E, Lin YL. Blocking of the alpha interferon-induced Jak-Stat signaling pathway by Japanese encephalitis virus infection. J Virol. 2004. 78:9285–9294.

113. Liu WJ, Wang XJ, Mokhonov VV, Shi PY, Randall R, Khromykh AA. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J Virol. 2005. 79:1934–1942.

114. Kalil AC, Devetten MP, Singh S, Lesiak B, Poage DP, Bargenquast K, et al. Use of interferon-alpha in patients with West Nile encephalitis: report of 2 cases. Clin Infect Dis. 2005. 40:764–766.

115. Rahal JJ, Anderson J, Rosenberg C, Reagan T, Thompson LL. Effect of interferon-alpha2b therapy on St. Louis viral meningoencephalitis: clinical and laboratory results of a pilot study. J Infect Dis. 2004. 190:1084–1087.

116. Lim TW, Yuan J, Liu Z, Qiu D, Sall A, Yang D. Nucleic-acid-based antiviral agents against positive single-stranded RNA viruses. Curr Opin Mol Ther. 2006. 8:104–107.
117. Voinnet O. Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet. 2005. 6:206–220.

118. Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001. 411:834–842.

119. Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009. 136:642–655.

120. Wu Z, Xue Y, Wang B, Du J, Jin Q. Broad-spectrum antiviral activity of RNA interference against four genotypes of Japanese encephalitis virus based on single microRNA polycistrons. PLoS One. 2011. 6:e26304.

121. Murakami M, Ota T, Nukuzuma S, Takegami T. Inhibitory effect of RNAi on Japanese encephalitis virus replication in vitro and in vivo. Microbiol Immunol. 2005. 49:1047–1056.

122. Geiss BJ, Pierson TC, Diamond MS. Actively replicating West Nile virus is resistant to cytoplasmic delivery of siRNA. Virol J. 2005. 2:53.

123. Geiss BJ, Stahla H, Hannah AM, Gari AM, Keenan SM. Focus on flaviviruses: current and future drug targets. Future Med Chem. 2009. 1:327–344.

124. Borowski P, Lang M, Haag A, Schmitz H, Choe J, Chen HM, et al. Characterization of imidazo[4,5-d] pyridazine nucleosides as modulators of unwinding reaction mediated by West Nile virus nucleoside triphosphatase/helicase: evidence for activity on the level of substrate and/or enzyme. Antimicrob Agents Chemother. 2002. 46:1231–1239.

125. Borowski P, Niebuhr A, Schmitz H, Hosmane RS, Bretner M, Siwecka MA, et al. NTPase/helicase of Flaviviridae: inhibitors and inhibition of the enzyme. Acta Biochim Pol. 2002. 49:597–614.

126. Johnston PA, Phillips J, Shun TY, Shinde S, Lazo JS, Huryn DM, et al. HTS identifies novel and specific uncompetitive inhibitors of the two-component NS2B-NS3 proteinase of West Nile virus. Assay Drug Dev Technol. 2007. 5:737–750.

127. Migliaccio G, Tomassini JE, Carroll SS, Tomei L, Altamura S, Bhat B, et al. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J Biol Chem. 2003. 278:49164–49170.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download