Abstract
Since 1994, several different inactivated rabies vaccines have been used to immunize domestic animals such as dogs, cats, and cattle in South Korea. The Korean Veterinary Authority has conducted safety and efficacy testes of inactivated vaccines using laboratory animals. In this study, we applied a molecular method to investigate the genetic characterization of the rabies virus (RABV) genes in six commercial inactivated rabies vaccines, and determined the efficiency of two extraction reagents (i.e., sodium citrate or isopropyl myristate) to separate the vaccine antigens from the antigen/adjuvant complexes. Six partial nucleocapsid (N: 181 bp) and five partial glycoprotein (G: 306 bp) genes were successfully amplified with specific primer sets, which demonstrated that sodium citrate is more efficient than isopropyl myristate in extracting viral RNA from inactivated gel vaccines. In addition, we identified the viral strain of the vaccine by analyzing the nucleotide sequences of the N and the G genes. The nucleotide similarity of the partial N and G genes ranged from 97.1 to 99.4% and from 91.8 to 100% among rabies vaccine strains, respectively, indicating that each manufacturer used different rabies virus strains to produce their vaccines. The molecular method used in this study could also be used to identify viral strains in other inactivated vaccines.
Go to : 
REFERENCES
1). Tordo N., Marianneau MP. Viruses and bats: rabies and Lyssavirus. Bull Mem Acad R Med Belg. 2009. 164:7–15.
2). World Health Organization (WHO). WHO Expert Committee on Rabies. World Health Organ Tech Rep Ser. 1992. 824:1–84.
3). Hyun BH., Lee KK., Kim IJ., Lee KW., Park HJ., Lee OS, et al. Molecular epidemiology of rabies virus isolates from South Korea. Virus Res. 2005. 114:113–25.

4). Kim CH., Lee CG., Yoon HC., Nam HM., Park CK., Lee JC, et al. Rabies, an emerging disease in Korea. J Vet Med B Infect Dis Vet Public Health. 2006. 53:111–5.

5). Park YJ., Shin MK., Kwon HM. Genetic characterization of rabies virus isolates in Korea. Virus Genes. 2005. 30:341–7.

6). Kwon YB., Kim YH., Lim YM. Studies on the production of rabies live vaccine. I. biological properties of the experimentally produced tissue culture attenuated live vaccine. Res Rep ORD. 1981. 23:125–35.
7). Hwang EK. Outbreak and control of rabies in animals in Korea. Korean J Vet Public Health. 1995. 19:281–93.
8). Lee JH., Lee MJ., Lee JB., Kim JS., Bae CS., Lee WC. Review of canine rabies prevalence under two different vaccination programmes in Korea. Vet Rec. 2001. 148:511–2.

9). Dreesen DW. Animal vaccine. Jackson AC & Wunner WH, editor. editors.Rabies. 2nd ed.London: Academic Press;2007. p. 517–27.
10). Animal and plant health inspection service (APHIS), Department of agriculture. 9 Code of Federal Regulations. 1-1-10th. Washington D. C: US government Printing Office. 2010. 741–3.
11). European pharmacopoeia. Strasbourg, European Directorate for the Quality of Medicines & Healthcare. 6th ed. 2007. 836–8.
12). Süliová J., Benísek Z., Svrcek S., Durove A., Ondrejka R. The effectiveness of inactivated, purified and concentrated experimental rabies vaccine for veterinary use: immunogenic activity. Vet Med (Praha). 1997. 42:51–6.
13). Yang J., Hooper DC., Wunner WH., Koprowski H., Dietzschold B., Fu ZF. The specificity of rabies virus RNA encapsidation by nucleoprotein. Virology. 1998. 242:107–17.

14). Hooper DC., Sauder C., Scott GS., Dietzschold B., Richt JA. Immunopathology and immunoprotection in CNS virus infections: mechanisms of virus clearance from the CNS. Curr Top Microbiol Immunol. 2002. 265:163–82.

15). Guyatt KJ., Twin J., Davis P., Holmes EC., Smith GA., Smith IL, et al. A molecular epidemiological study of Australian bat lyssavirus. J Gen Virol. 2003. 84:485–96.

16). Morimoto K., Hooper DC., Spitsin S., Koprowski H., Dietzschold B. Pathogenicity of different rabies virus variants inversely correlates with apoptosis and rabies virus glycoprotein expression in infected primary neuron cultures. J Virol. 1999. 73:510–8.

17). Choi JG., Lee YJ., Kim JY., Kim YH., Paek MR., Yang DK, et al. Molecular identification of the vaccine strain from the inactivated oil emulsion H9N2 low pathogenic avian influenza vaccine. J Vet Sci. 2010. 11:161–3.

18). Maas R., van Diepen M., Komen M., Oei H., Claassen I. Antigen content of inactivated Newcastle disease oil emulsion vaccines as an in vitro indicator of potency. Dev Biol (Basel). 2002. 111:313–8.
Go to : 
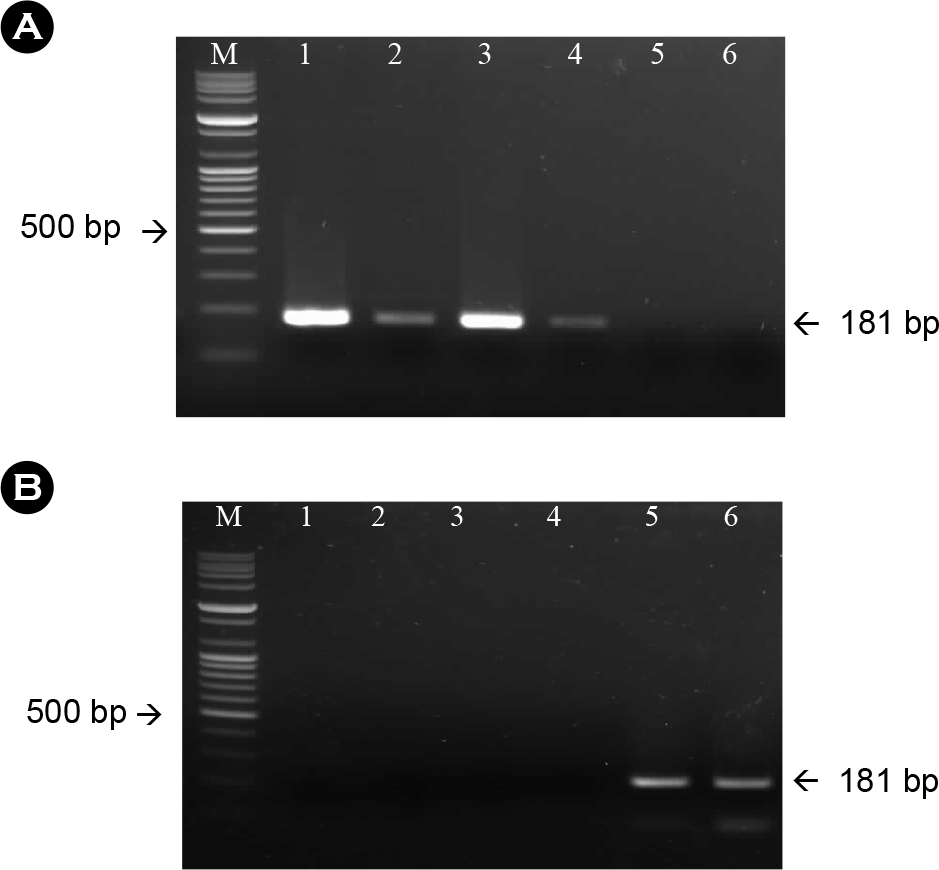 | Figure 1.Amplification of the N gene from the rabies virus using RNA extracted from sodium citrate-treated antigen (A) and isopropyl myristate-treated antigen (B) with specific primer sets for the N gene. The expected size was 181 bp. M: 100-bp DNA ladder; lane 1-6: company A, B, C, D, E, F, respectively. |
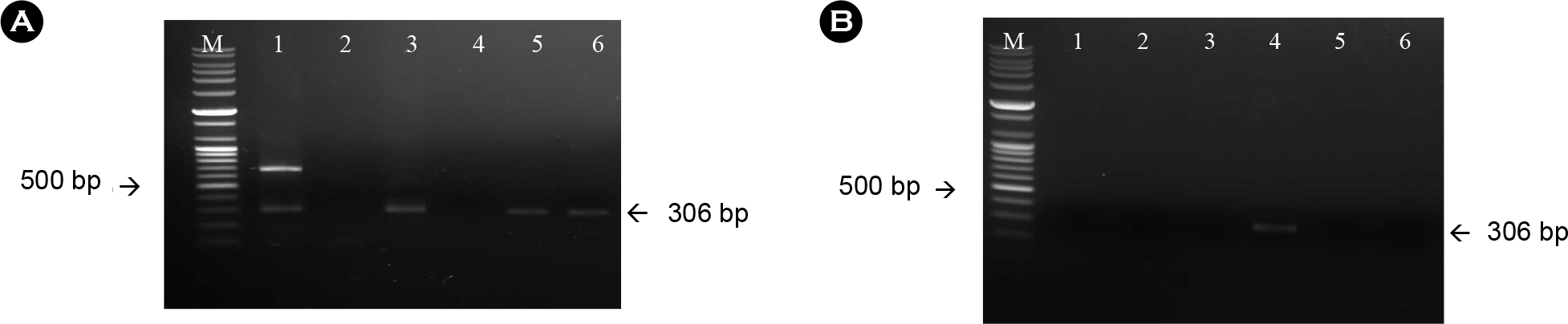 | Figure 2.Amplification of the G gene from the rabies virus using RNA extracted from sodium citrate-treated antigen (A) and isopropyl myristate-treated antigen (B) with specific primer sets for the G gene. The expected size was 181 bp. M: 100-bp DNA ladder; lane 1: company A; lane 2: company C; lane 3: company B; lane 4~6: D, E and F, respectively. |
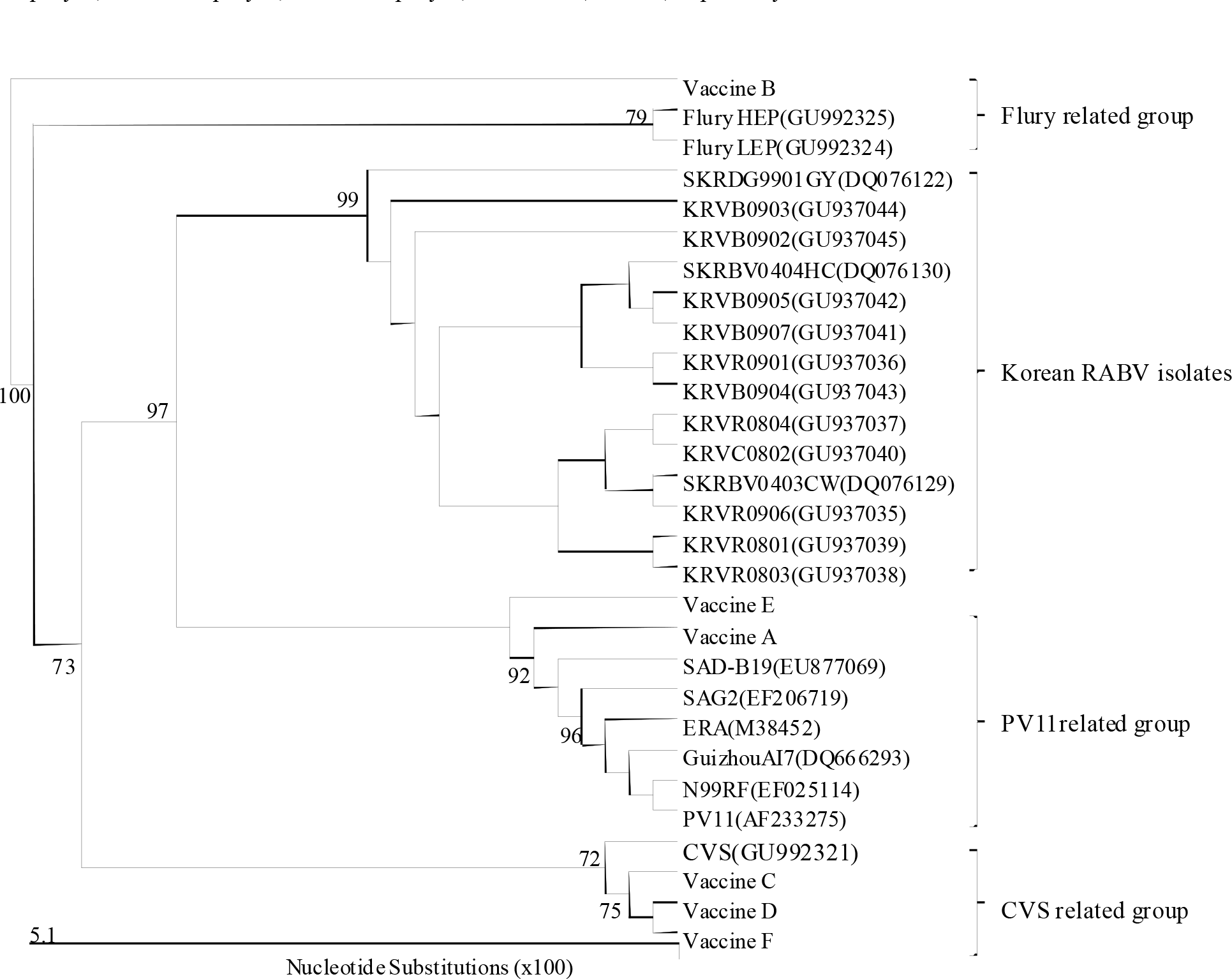 | Figure 3.Phylogenetic analysis based on the partial N-gene nucleotide sequences of the vaccine strains and other sequences obtained from the GenBank database. Numbers at each key node indicate the degree of bootstrap support and only those with >70% support are shown. |
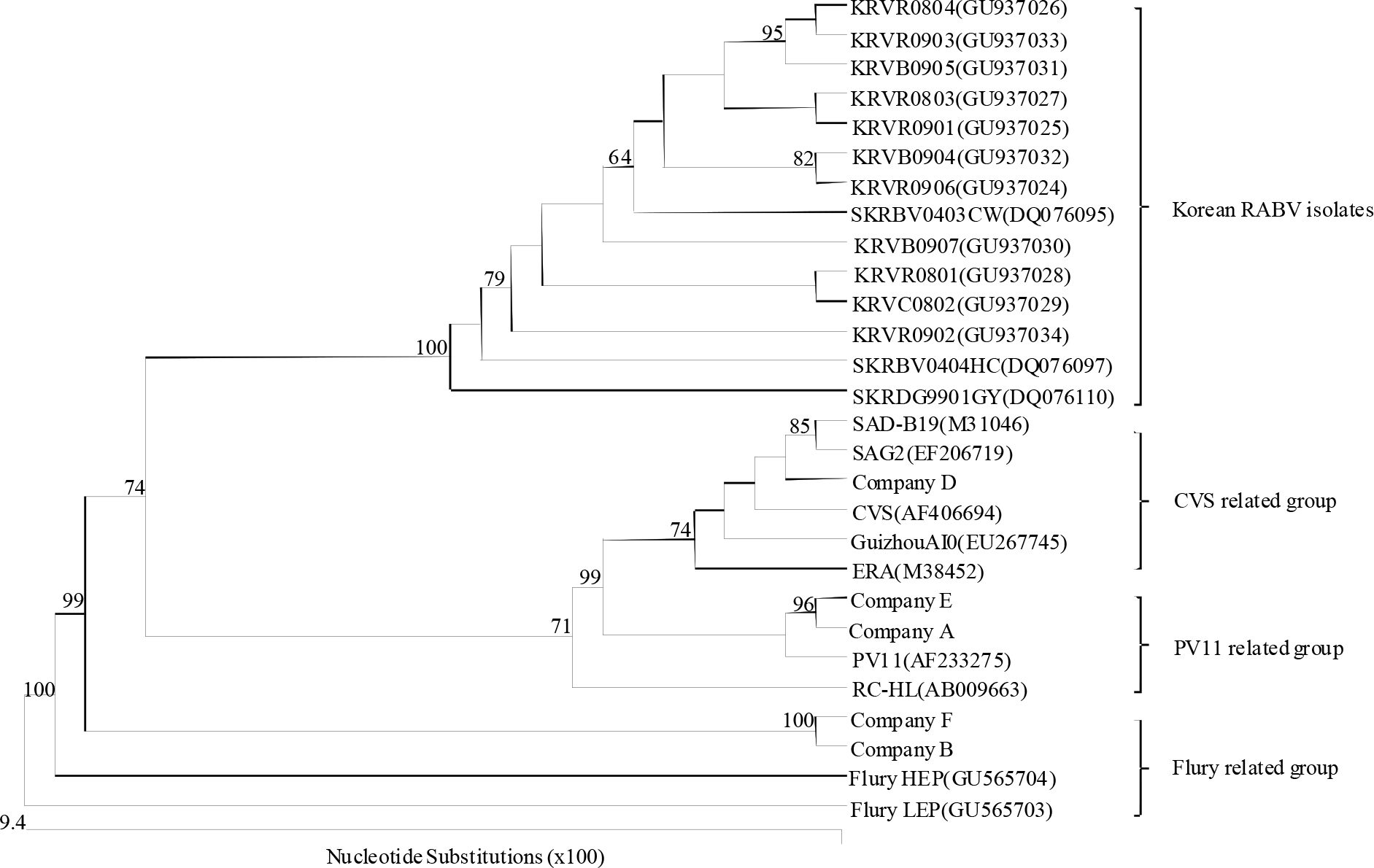 | Figure 4.Phylogenetic analysis based on the partial G gene nucleotide sequences of the vaccine strains and other sequences obtained from the GenBank database. Numbers at each key node indicate the degree of bootstrap support and only those with >70% support are shown. |
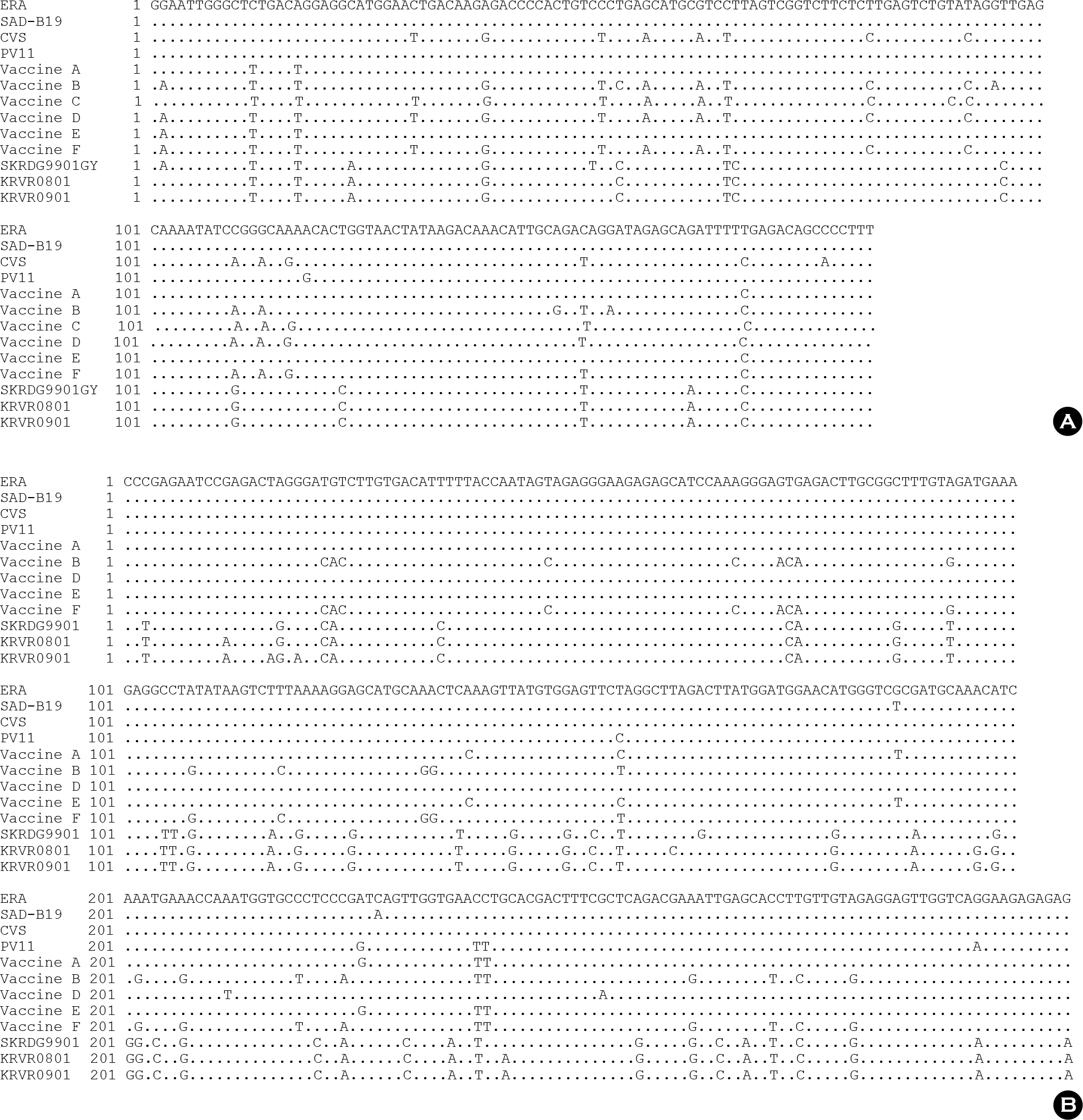 | Figure 5.Comparison of the nucleotide sequences of nucleocapsid (A) and glycoprotein (B) genes of the vaccine strains and representative Korean RABV isolates. Dots indicate nucleotides agreeing with the first line sequence. |
Table 1.
List of the oligonucleotide primers used for RT-PCR of the rabies vaccine
| Primer | Nucleotide sequences (5′-3′) | Nucleotide positiona | Sense | RABV gene | Size of amplicon (bp) |
|---|---|---|---|---|---|
| RVNDF | GRA ATT GGG CTT TGA CTG GA | 353~372 | + | N | 181 |
| RFNDR | AAA GGG GCT GTC TCG AAA AT | 514~533 | – | ||
| RVGDF | CCC GAG AAT CCG AGA CTA | 595~612 | + | G | 306 |
| RFGDR | CTC TCT CTT CCT GAC CAA CTC | 880~900 | – |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download