Abstract
The genus Legionella is common in aquatic environments. Some species of Legionella are recognized as potential opportunistic pathogens for human, notably Legionella pneumophila that causes Legionellosis. During the summer season between June and August in 2010, we isolated 61 L. pneumophila from the bath facilities of public establishments of 25 wards in Seoul. The existence of 16S rRNA and mip gene of L. pneumophila was confirmed in the genome of the isolated strains by PCR. Among the 61 strains of L. pneumophila, thirty three isolates belong to serogroup 1 (54.1%), 13 isolates were serogroup 6 (21.3%), 9 isolates were serogroup 5 (14.8%), 3 isolates were serogroup 3 (4.9%), and 3 isolates were identified in serogroup 2 (4.9%). On pulsed-field gel electrophoresis (PFGE) analysis using SfiI, genetic types of L. pneumophila were classified into 8 (A to H) patterns by the band similarity with excess of 65%. Our results suggest the existence of serological and genetic diversity among the L. pneumophila isolates.
Go to : 
REFERENCES
1). Fields BS, Benson RF, Besser RE. Legionella and Legionnaires' disease: 25 Years of investigation. Clin Microbiol Rev. 2002; 15:506–26.
2). Nintasen R, Utrarachkij F, Siripanichgon K, Bhumiratana A, Suzuki Y, Suthienkul O. Enhancement of Legionella pneumophila culture isolation from microenvironments by macrophage infectivity potentiator (mip) gene-specific nested polymerase chain reaction. Microbiol Immunol. 2007; 51:777–85.
3). Amemura-Maekawa J, Kura F, Chang B, Watanabe H. Legionella pneumophila serogroup 1 isolates from cooling towers in Japan form a distinct genetic cluster. Microbiol Immunol. 2005; 49:1027–33.
4). Cooper IR, White J, Mahenthiralingam E, Hanlon GW. Long-term persistence of a single Legionella pneumophila strain possessing the mip gene in a municipal shower despite repeated cycles of chlorination. J Hosp Infect. 2008; 70:154–9.
5). Köhler R, Fanghänel J, König B, Lüneberg E, Frosch M, Rahfeld JU, et al. Biochemical and functional analyses of the mip protein: Influence of the N-terminal half and of peptidylprolyl isomerase activity on the virulence of Legionella pneumophila. Infect Immun. 2003; 71:4389–97.
6). Cianciotto NP, Eisenstein BI, Mody CH, Engleberg NC. A mutation in the mip gene results in an attenuation of Legionella pneumophila virulence. J Infect Dis. 1990; 162:121–6.
7). Schoonmaker D, Heimberger T, Birkhead G. Comparison of ribotyping and restriction enzyme analysis using pulsed-field gel electrophoresis for distinguishing Legionella pneumophila isolates obtained during a nosocomial outbreak. J Clin Microbiol. 1992; 30:1491–8.
8). Benson RF, Fields BS. Classification of the genus Legionella. Semin Respir Infect. 1998; 13:90–9.
9). Hsu BM, Chen CH, Wan MT, Cheng HW. Legionella prevalence in hot spring recreation areas of Taiwan. Water Res. 2006; 40:3267–73.
10). Kim SH, Kim MJ, Kee HY, Kim TS, Seo JJ, Kim ES, et al. Surveillance of Legionella contamination on water supply systems of public utilizing facilities in Gwangju, Korea. J Bacteriol Virol. 2010; 40:19–28.
11). Whang KH, Hwang YO, Kim EJ, Jung JH, Cho NJ. Distribution and serological characteristics of Legionella spp. isolated from cooling tower waters of public establishments in Seoul. Kor J Env Hlth Soc. 1999; 25:20–3.
12). Kim JA, Jung JH, Kim SJ, Jin YH, Oh YH, Han GY. Molecular epidemiological relationship of the pathogenicity of Legionella spp. isolated from water systems in Seoul. Kor J Microbiol. 2009; 45:126–32.
13). Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995; 33:2233–9.

14). Kim KY, Kim YS, Song JC, Lee SJ, Kim SU, Choi TY, et al. Comparison of method for identification and the effects on Legionella pneumophila of the cooling towers in Seoul. J Korean Occup Environ. 2003; 13:234–52.
15). Park EH, Kim MH, Kim JA, Han NS, Lee JH, Min SG, et al. Molecular typing of Legionella pneumophila isolated in Busan, using PFGE. J Life Sci. 2005; 15:161–8.
Go to : 
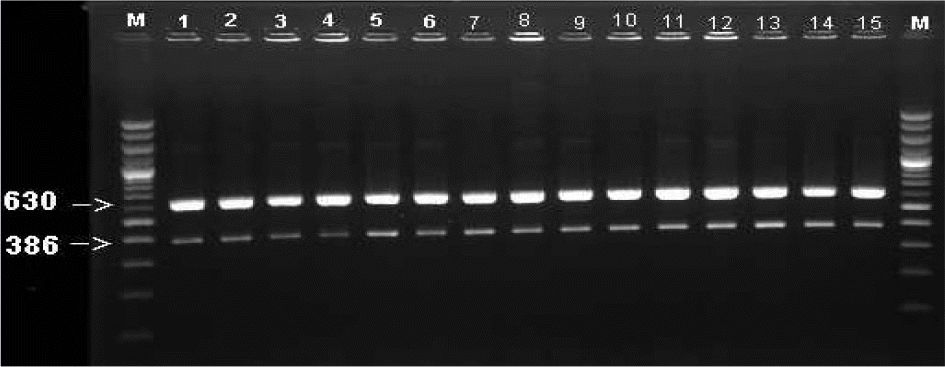 | Figure 1.Gel electrophoresis of PCR products of mip gene (630 bp) and 16S rRNA (386 bp) on 1.5% agarose gel. M was 100 bp DNA ladder. All lanes (1∼15) were showed bands of correct size for L. pneumophila.
|
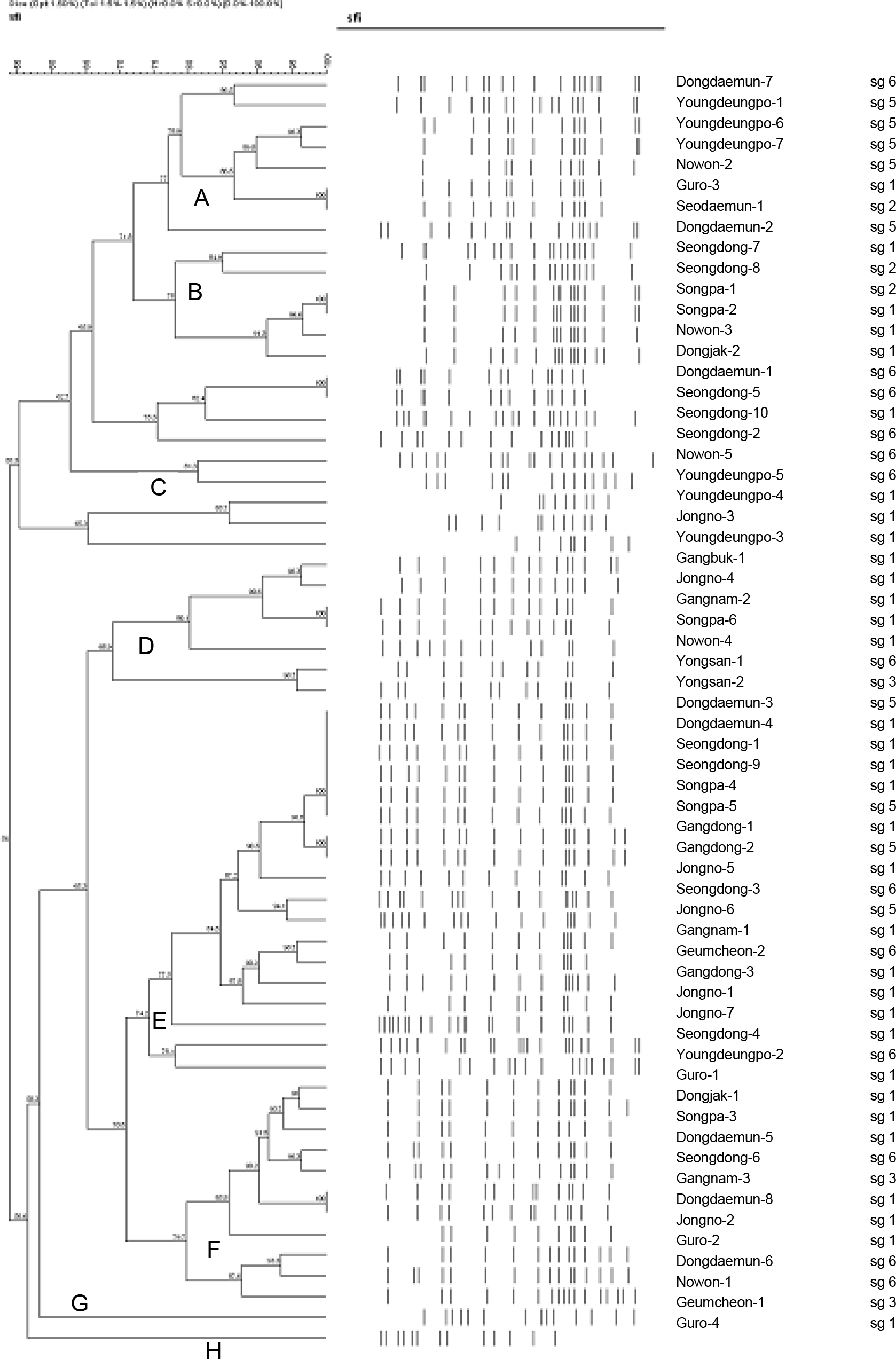 | Figure 2.Dendrogram showing the clustering of PFGE patterns of isolated strains of L. pneumophila after SfiI digestion. |
Table 1.
Primers used in this study
| Primer | Sequence (5′ → 3′) | Product size |
|---|---|---|
| 16S rRNA | F: AGGGTTGATAGGTTAAGAGG | 386 bp |
| R: CAACAGCTAGTTGACATGG | ||
| mip | F: GGTGACTGCGGCTGTTATGG | 630 bp |
| R: GGCCAATAGGTCCGCCAACG |
Table 2.
Serotyping of L. pneumophila strains isolated from bath facilities of public establishments in Seoul
| Species | Serogroup | Number of isolates (%) |
|---|---|---|
| L. pneumophila | 1 | 33 (54.1%) |
| 6 | 13 (21.3%) | |
| 5 | 9 (14.8%) | |
| 3 | 3 (4.9%) | |
| 2 | 3 (4.9%) | |
| 4 | 0 | |
| Total | 61 |
Table 3.
Categories of PFGE and serogroup (sg) pattern of L. pneumophila strains isolated from bath facilities of public establishments in Seoul




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download