Abstract
Resistance to mupirocin in methicillin-resistant Staphylococcus aureus (MRSA) have increased with wide use of mupirocin in many countries, but the prevalence in Korea is not well-known. The aim of this study was to determine the prevalence, antimicrobial susceptibility, and clonality of mupirocin-resistant (MUP-R) isolates from three Korean hospitals. A total of 175 MRSA isolates were collected from three university hospitals in 2009–2010. Antimicrobial susceptibility was tested by the disk diffusion and the agar dilution methods. femA, mecA and mupA genes were detected by polymerase chain reactions. Pulsed-filed gel electrophoresis (PFGE) pattern of genomic DNA was determined after digestion with SmaI. Overall, 12 among the 175 MRSA isolates were resistant to mupirocin, with prevalence ranging from 0 to 10% depending on hospitals. Three high-level (HL) and nine low-level (LL) MUR-R isolates were obtained from two hospitals. All MUP-R isolates were susceptible to rifampin and vancomycin, but were resistant to ciprofloxacin, clindamycin, and erythromycin. Eight LL and one HL MUP-R isolates were also resistant to fusidic acid. PFGE analysis showed three HL MUP-R isolates belonged to arbitrary cluster 3, 5 and 6 with 60∼90% similarity compared to LL MUP-R isolates. In conclusion, the HL resistance to mupirocin was detected in two hospitals, but HL MUP-R isolates were clonally not related.
REFERENCES
1). Styers D, Sheehan DJ, Hogan P, Sahm DF. Laboratory-based surveillance of current antimicrobial resistance patterns and trends among Staphylococcus aureus: 2005 status in the United States. Ann Clin Microbiol Antimicrob. 2006; 5:2.

2). Yoo JI, Shin ES, Cha JO, Lee JK, Jung YH, Lee KM, et al. Clonal dissemination and mupA gene polymorphism of mupirocin-resistant Staphylococcus aureus isolates from long-term-care facilities in South Korea. Antimicrob Agents Chemother. 2006; 50:365–7.
3). Kauffman CA, Terpenning MS, He X, Zarins LT, Ramsey MA, Jorgensen KA, et al. Attempts to eradicate methicillin-resistant Staphylococcus aureus from a long-term-care facility with the use of mupirocin ointment. Am J Med. 1993; 94:371–8.
4). Ramsey MA, Bradley SF, Kauffman CA, Morton TM. Identification of chromosomal location of mupA gene, encoding low-level mupirocin resistance in Staphylococcal isolates. Antimicrob Agents Chemother. 1996; 40:2820–3.
5). Cookson BD. The emergence mupirocin resistance: a challenge to infection control and antibiotic prescribing practice. J Antimicrob Chemother. 1998; 41:11–8.
6). No authors listed. Mupirocin-resistant Staphylococcus aureus. Lancet. 1987; 2:387–8.
7). Yun HJ, Lee SW, Yoon GM, Kim SY, Choi S, Lee YS, et al. Prevalence and mechanisms of low- and high-level mupirocin resistance in staphylococci isolated from a Korean hospital. J Antimicrob Chemother. 2003; 51:619–23.

8). Layton MC, Perez M, Heald P, Patterson JE. An outbreak of mupirocin-resistant Staphylococcus aureus on a dermatology ward associated with an environmental reservoir. Infect Control Hosp Epidemiol. 1993; 14:369–75.
9). Layton MC, Patterson JE. Mupirocin resistance among consecutive isolates of oxacillin-resistant and borderline oxacillin-resistant Staphylococcus aureus at a university hospital. Antimicrob Agents Chemother. 1994; 38:1664–7.
10). Gilbart J, Perry CR, Slocombe B. High-level mupirocin resistance in Staphylococcus aureus: evidence for two distinct isoleucyl-tRNA synthetases. Antimicrob agents chemother. 1993; 37:32–8.
11). Janssen DA, Zarins LT, Schaberg DR, Bradley SF, Terpenning MS, Kauffman CA. Detection and characterization of mupirocin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1993; 37:2003–6.
12). Pérez-Roth E, López-Aguilar C, Alcoba-Florez J, Méndez-Alvarez S. High-level mupirocin resistance within methicillin-resistant Staphylococcus aureus pandemic lineages. Antimicrob Agents Chemother. 2006; 50:3207–11.
13). Decousser JW, Pina P, Ghnassia JC, Bedos JP, Allouch PY. First report of clinical and microbiological failure in the eradication of glycopeptide-intermediate methicillin-resistant Staphylococcus aureus carriage by mupirocin. Eur J Clin Microbiol Infec Dis. 2003; 22:318–9.
14). Yokoyama T. Study on mec gene in methicillin-resistant Staphylococci. Kansenshogaku Zasshi. 1993; 67:1203–10.
15). Lee HK, Lee EJ, Pahk YJ, Kim BK, Kang MW, Shim SI. Relationship between the level of methicillin resistance and mecA, mecI, femA, genes in staphylococci. Korean J Infect Dis. 1998; 30:36–44.
16). Kim SM, Lee DC, Park SD, Kim BS, Kim JK, Choi MR, et al. Genotype, coagulase type and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolated from dermatology patients and healthy individuals in Korea. J Bacteriol Virol. 2009; 39:307–16.
17). Cockerill FR, Wikler MA, Bush K, Dudley MN, Eliopoulos GM, Hardy DJ, et al. Performance standards for antimicrobial susceptibility testing twenty-first informational supplement. M100-S21. Clinical and Laboratory Standards Institute;Wayne PA: 2011. P.p. 68–80.
18). Finlay JE, Miller LA, Poupard JA. Interpretive criteria for testing susceptibility of staphylococci to mupirocin. Antimicrob Agents Chemother. 1997; 41:1137–9.

19). Norazah A, Koh YT, Ghani Kamel A, Alias R, Lim VK. Mupirocin resistance among Malaysian isolates of methicillin-resistant Staphylococcus aureus. Int J Antimicrob Agents. 2001; 17:411–4.
20). Hodgson JE, Curnock SP, Dyke KG, Morris R, Sylvester DR, Gross MS. Molecular characterization of the gene encoding high level mupirocin resistance in Staphylococcus aureus J2870. Antimicrob Agents Chemother. 1994; 38:1205–8.
21). Udo EE, Jacob LE, Mathew B. Genetic analysis of methicillin-resistant Staphylococcus aureus expressing high- and low-level mupirocin resistance. J Med Microbiol. 2001; 50:909–15.
22). Murchan S, Kaufmann ME, Deplano A, de Ryck R, Struelens M, Zinn CE, et al. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol. 2003; 41:1574–85.
23). Graves SF, Kobayashi SD, DeLeo FR. Community-associated methicillin-resistant Staphylococcus aureus immune evasion and virulence. J Mol Med. 2010; 88:109–14.
24). Shittu AO, Udo EE, Lin J. Phenotypic and molecular characterization of Staphylococcus aureus isolates expressing low- and high-level mupirocin resistance in Nigeria and south Africa. BMC Infect Dis. 2009; 9:10.

25). Fujimura S, Watanabe A. Survey of high- and low-level mupirocin-resistant strains of methicillin-resistant Staphylococcus aureus in 15 Japanese hospitals. Chemotherapy. 2003; 49:36–8.
26). Walker ES, Vasquez JE, Dula R, Bullock H, Sarubbi FA. Mupirocin-Resistant, methicillin-resistant Staphylococcus aureus: does mupirocin remain effective? Infect Control Hosp Epidemiol. 2003; 24:342–6.
27). Lee AJ, Suh HS, Jeon CH, Kim SG. Prevalence and clinical charateristics of mupirocin-resistant Staphylococcus aureus. Korean J Clin Microbiol. 2011; 14:18–23.
28). Fujimura S, Watanabe A, Beighton D. Charaterization of the mupA gene in strains of methicillin-resistant Staphylococcus aureus with a low level of resistance to mupirocin. Antimicrob Agents Chemother. 2001; 45:641–2.
29). Simor AE, Stuart TL, Louie L, Watt C, Ofner-Agostini M, Gravel D, et al. Mupirocin-resistant, methicillin-resistant Staphylococcus aureus strains in Canadian hospitals. Antimicrob Agents Chemother. 2007; 51:3880–6.
30). Lim KT, Hanifah YA, Mohd Yusof MY, Thong KL. Prevalence of mupirocin resistance methicillin-resistant Staphylococcus aureus strains isolated from a Malaysian hospital. Jpn J Infect Dis. 2010; 63:286–9.
31). Chaves F, García-Martínez J, de Miguel S, Otero JR. Molecular characterization of resistance to mupirocin in methicillin-susceptible and -resistant isolates of Staphylococcus aureus from nasal samples. J Clin Microbiol. 2004; 42:822–4.
Figure 1.
A dendrogram generated with Fingerprinting II informatix software showing the PFGE types (1–6) of SmaI-restricted chromosome DNA of 12 mupirocin-resistant MRSA isolates. No. 03, 07, and 08 isolates with high-level resistance to mupirocin; No. 01∼02, 4∼6, 9∼12 isolates with low-level resistance to mupirocin.
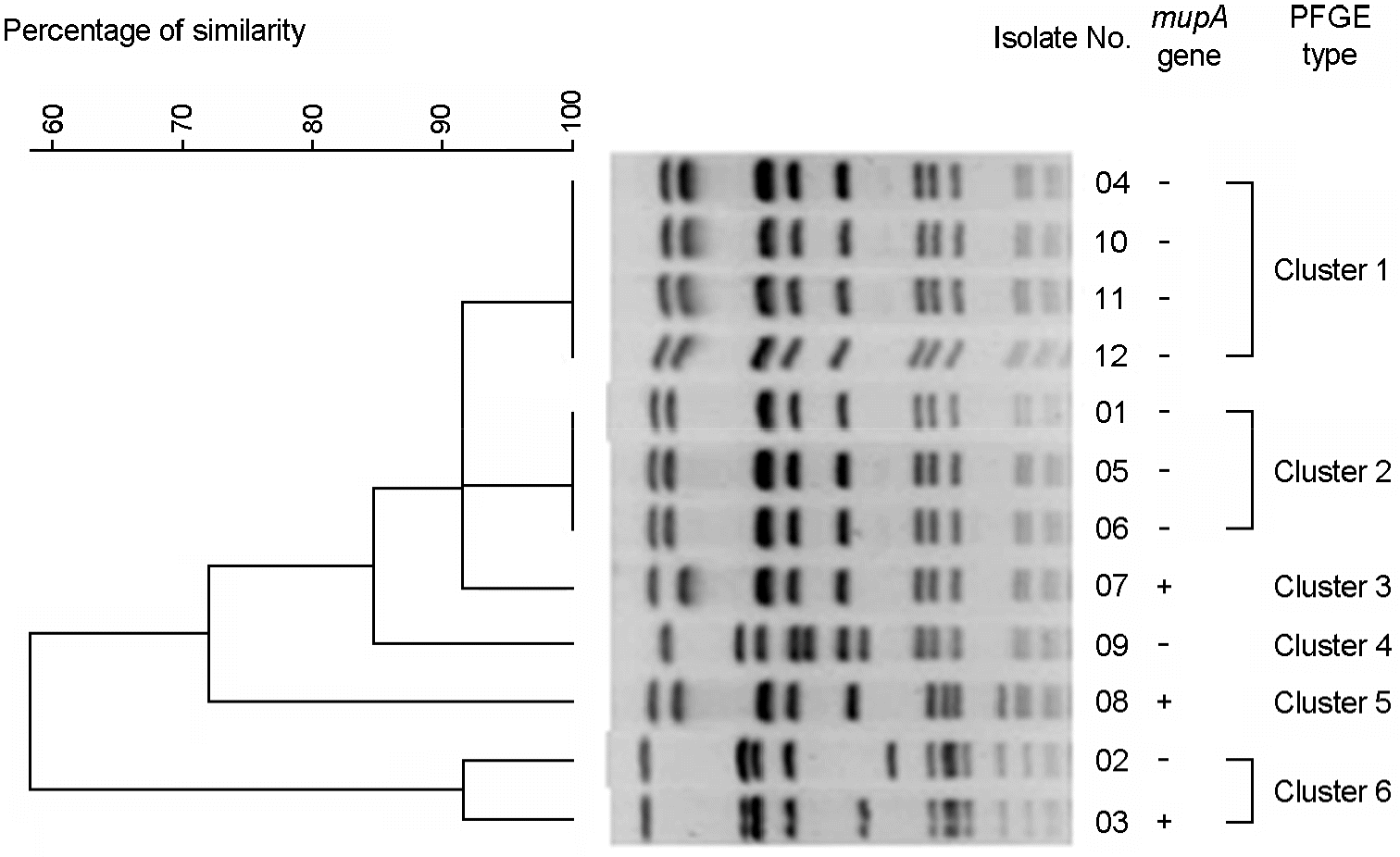
Table 1.
Mupirocin-resistant MRSA isolates detected in three Korean hospitals
Table 2.
Antimicrobial susceptibility of high- and low-level mupirocin-resistant MRSA isolates determined by disk diffusion
| Antimicrobial agents |
HL MUP-R MRSAa (n = 3)b |
LL MUP-R MRSA (n = 9) | ||||
|---|---|---|---|---|---|---|
| Sc | I | R | S | I | R | |
| Amikacin | 0 | 33 | 67 | 33 | 11 | 56 |
| Gentamicin | 0 | 0 | 100 | 22 | 0 | 78 |
| Tobramycin | 0 | 0 | 100 | 22 | 0 | 78 |
| Chloramphenicol | 100 | 0 | 0 | 100 | 0 | 0 |
| Ciprofloxacin | 0 | 0 | 100 | 0 | 0 | 100 |
| Clindamycin | 0 | 0 | 100 | 0 | 0 | 100 |
| Erythromycin | 0 | 0 | 100 | 0 | 0 | 100 |
| Mupirocin | 0 | 0 | 100 | 0 | 0 | 100 |
| Tetracycline | 33 | 0 | 67 | 11 | 0 | 89 |
| Vancomycin | 100 | 0 | 0 | 100 | 0 | 0 |
Table 3.
MICs of high- and low-level mupirocin-resistant MRSA isolates against antimicrobial agents




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download