Abstract
Swine is a common source of Campylobacter coli human gastroenteritis, for the treatment of which erythromycin and fluoroquinolones are recommended. The prevalence of antimicrobial-resistant C. coli differs significantly depending on countries. We investigated the prevalence of C. coli in swine from a farm in Buan-gun, Korea in 2010, and determined antimicrobial susceptibility of the isolates. Rectal swab specimens were used to inoculate Campylobacter Preston media and incubated microaerophilically at 42°C for 48 h. The species were identified by phenotypic tests and by detecting hipO and glyA genes. PCR was used to detect mutations of A2074C in 23S rRNA gene, and quinolone resistance-determining region (QRDR) of gyrA, which are associated with high level resistance to erythromycin, and with ciprofloxacin, respectively. Antimicrobial susceptibility was determined by the disk diffusion and agar dilution tests. Of the 100 specimens, 55 (55%) yielded C. coli, and 23 of them (41.8%) had A2074G mutation. A2074G mutated isolates showed the lowest MIC90 of imipenem, while those of ampicillin and clindamycin were relatively low. The majority of both A2074G mutation-positive and -negative isolate were susceptible to ampicillin, cefotaxime, and chloramphenicol. All isolates were resistant to ciprofloxacin, and had mutation in QRDR of gyrA. In conclusion, C. coli was detected in 55% of swine, and A2074G mutation was detected in 41.8% of the isolates. All isolates had gyrA mutation-mediated ciprofloxacin resistance.
Go to : 
REFERENCES
1). Friedman J., Neimann J., Wegener HC., Tauxe RV. Epidemiology of Campylobacter jejuni infection in the united statesand other industrialized nations. Nackamkin I, Blaser MJ, editors. editors,. Campylobacter. 2nd ed.Washington D.C.: ASM Press;2000. p. 121–38.
2). Aarestrup FM., Engberg J. Antimicrobial resistance of thermophilic Campylobacter. Vet Res. 2001. 32:311–21.
3). Aarestrup FM., Nielsen EM., Madsen M., Engberg J. Antimicrobial susceptibility patterns of thermophilic Campylobacter spp. from humans, pigs, cattle, and broilers in Denmark. Antimicrob Agents Chemother. 1997. 41:2244–50.
4). Allos BM. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis. 2001. 32:1201–6.
5). Harrow SA., Gilpin BJ., Klena JD. Characterization of erythromycin resistance in Campylobacter coli and Campylobacter jejuni isolated from pig offal in New Zealand. J Appl Microbiol. 2004. 97:141–8.
6). Gibreel A., Taylor DE. Macrolide resistance in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother. 2006. 58:243–55.
7). Vanhoof R., Vanderlinden MP., Dierickx R., Lauwers S., Yourassowsky E., Butzler JP. Susceptibility of Campylobacter fetus subsp. jejuni to twenty nine antimicrobial agents. Antimicrob Agents Chemother. 1978. 14:553–6.
8). Li CC., Chiu CH., Wu JL., Huang YC., Lin TY. Antimicrobial susceptibilities of Campylobacter jejuni and coli by using E-test in Taiwan. Scand J Infect Dis. 1998. 30:39–42.
9). Saenz Y., Zarazaga M., Lantero M., Gastanares MJ., Baquero F., Torres C. Antibiotic resistance in Campylobacter strains isolated from animals, foods, and humans in Spain in 1997~1998. Antimicrob Agents Chemother. 2000. 44:267–71.
10). Luber P., Wagner J., Hahn H., Bartelt E. Antimicrobial resistance in Campylobacter jejuni and Campylobacter coli strains isolated in 1991 and 2001~2002 from poultry and humans in Berlin, Germany. Antimicrob Agents Chemother. 2003. 47:3825–30.
11). Pezzotti G., Serafin A., Luzzi I., Mioni R., Milan M., Perin R. Occurrence and resistance to antibiotics of Campylobacter jejuni and Campylobacter coli in animals and meat in northeastern Italy. Int J Food Microbiol. 2003. 82:281–7.
12). Gupta A., Nelson JM., Barrett TJ., Tauxe RV., Rossiter SP., Friedman CR, et al. Antimicrobial resistance among Campylobacter strains, United States, 1997~2001. Emerg Infect Dis. 2004. 10:1102–9.
13). Chuma T., Ikeda T., Maeda T., Niwa H., Okamoto K. Antimicrobial susceptibilities of Campylobacter strains isolated from broilers in the southern part of Japan from 1995 to 1999. J Vet Med Sci. 2001. 63:1027–9.
14). Van Looveren M., Daube G., De Zutter L., Dumont JM., Lammens C., Wijdooghe M, et al. Antimicrobial susceptibilities of Campylobacter strains isolated from food animals in Belgium. J Antimicrob Chemother. 2001. 48:235–40.
15). Alonso R., Mateo E., Churruca E., Martinez I., Girbau C., Fernández-Astorga A. MAMA-PCR assay for the detection of point mutations associated with high-level erythromycin resistance in Campylobacter jejuni and Campylobacter coli strains. J Microbiol Methods. 2005. 63:99–103.
16). Niwa H., Chuma T., Okamoto K., Itoh K. Rapid detection of mutations associated with resistance to erythromycin in Campylobacter jejuni/coli by PCR and line probe assay. Int J Antimicrob Agents. 2001. 18:359–64.
17). Vacher S., Ménard A., Bernard E., Mégraud F. PCR-restriction fragment length polymorphism analysis for detection of point mutations associated with macrolide resistance in Campylobacter spp. Antimicrob Agents Chemother. 2003. 47:1125–8.
18). Payot S., Avrain L., Magras C., Praud K., Cloeckaert A., Chaslus-Dancla E. Relative contribution of target gene mutation and efflux to fluoroquinolone and erythromycin resistance, in French poultry and pig isolates of Campylobacter coli. Int J Antimicrob Agents. 2004. 23:468–72.
19). Vacher S., Menard A., Bernard E., Santos A., Megraud F. Detection of mutations associated with macrolide resistance in thermophilic Campylobacter spp. by real-time PCR. Microb Drug Resist. 2005. 11:40–7.
20). Perez-Boto D., Lopez-Portoles JA., Simon C., Valdezate S., Echeita MA. Study of the molecular mechanisms involved in high-level macrolide resistance of Spanish Campylobacter jejuni and Campylobacter coli strains. J Antimicrob Chemother. 2010. 65:2083–8.
21). Shin E., Lee Y. Characterization of erythromycin-resistant porcine isolates of Campylobacter coli. Microb Drug Resist. 2010. 16:231–9.
22). Fitzgerald C., Nachamkin I. Campylobacter and Arcobacter. Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA, editors. editors.Manual of clinical microbiology. 9th ed.Washington, D.C.: ASM Press;2007. p. 933–62.
23). Wang G., Clark CG., Taylor TM., Pucknell C., Barton C., Price L, et al. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis and C. fetus subsp. fetus. J Clin Microbiol. 2002. 40:4744–7.
24). Zirnstein G., Li Y., Swaminathan B., Angulo F. Ciprofloxacin resistance in Campylobacter jejuni isolates: detection of gyrA resistance mutations by mismatch amplification mutation assay PCR and DNA sequence analysis. J Clin Microbiol. 1999. 37:3276–80.
25). CLSI. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline. CLSI Document Clincal and Laboratory Standards Institute M45-A. Wayne PA. 2006. 26:16–7.
26). Guévremont E., Nadeau E., Sirois M., Quessy S. Antimicrobial susceptibilities of thermophilic Campylobacter from humans, swine, and chicken broilers. Can J Vet Res. 2006. 70:81–6.
27). Nielsen EM., Engberg J., Madsen M. Distribution of serotypes of Campylobacter jejuni and C. coli from Danish patients, poultry, cattle and swine. FEMS Immunol Med Microbiol. 1997. 19:47–56.
28). Alter T., Gaull F., Kasimir S., Gürtler M., Fehlhaber K. Distribution and genetic characterization of porcine Campylobacter coli isolates. Berl Munch Tierarztl Wochenschr. 2005. 118:214–9.
29). Dassanayake RP., Zhou Y., Hinkley S., Stryker CJ., Plauche G., Borda JT, et al. Characterization of cytolethal distending toxin of Campylobacter species isolated from captive macaque monkeys. J Clin Microbiol. 2005. 43:641–9.
30). Swartz MN. Human diseases caused by foodborne pathogens of animal origin. Clin Infect Dis. 2002. 34:S111–22.

31). Davies P., Morrow M., Funk J., Deen J. Erythromycin resistance of Campylobacter isolates from pigs. Vet Rec. 1996. 139:244.
32). Moore JE., Madden RH., Kerr JR., Wilson TS., Murphy PG. Erythromycin-resistant thermophilic Campylobacter species isolated from pigs. Vet Rec. 1996. 138:306–7.
33). Ishihara K., Kira T., Ogikubo K., Morioka A., Kojima A., Kijima-Tanaka M, et al. Antimicrobial susceptibilities of Campylobacter isolated from food-producing animals on farms (1999~2001):results from the Japanese Veterinary Antimicrobial Resistance Monitoring Program. Int J Antimicrob Agents. 2004. 24:261–7.
34). Kim SM., Kim EC., Choi MR., So HA., Shim ES., Kim ES, et al. Cytolethal distending toxin production, genotypes and antimicrobial susceptibility of Campylobacter jejuni isolates from diarrhea patients and chickens. J Bacteriol Virol. 2008. 38:207–19.
35). Boonmar S., Sangsuk L., Suthivarakom K., Padungtod P., Morita Y. Serotypes and antimicrobial resistance of Campylobacter jejuni isolated from humans and animals in Thailand. Southeast Asian J Trop Med Public Health. 2005. 36:130–4.
36). Segreti J., Nelson JA., Goodman LJ., Kaplan RL., Trenholme GM. In vitro activities of lomefloxacin and temafloxacin against pathogens causing diarrhea. Antimicrob Agents Chemother. 1989. 33:1385–7.
37). Goodman LJ., Trenholme GM., Kaplan RL., Segreti J., Hines D., Petrak R, et al. Empiric antimicrobial therapy of domestically acquired acute diarrhea in urban adults. Arch Intern Med. 1990. 150:541–6.

38). Zaman R. Campylobacter enteritis in Saudi Arabia. Epidemiol Infect. 1992. 108:51–8.
Go to : 
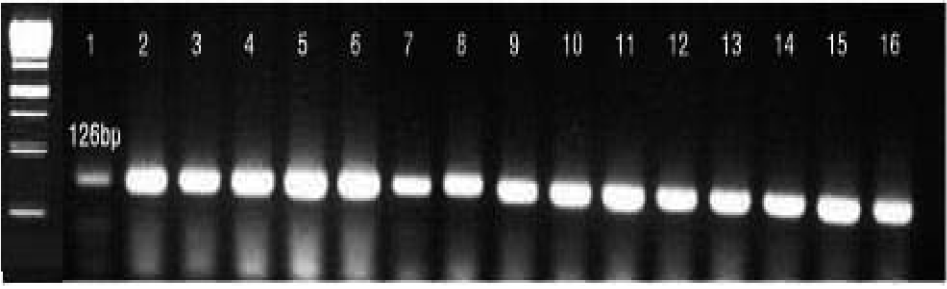 | Figure 1.Agarose gel electrophoresis of PCR-generated amplicons by using primer pairs for glyA gene for identification of C. coli. Lane M: size marker, lane 1 to 15: isolate no. C01-1 to C01-15, lane 16: C. coli ATCC3359. |
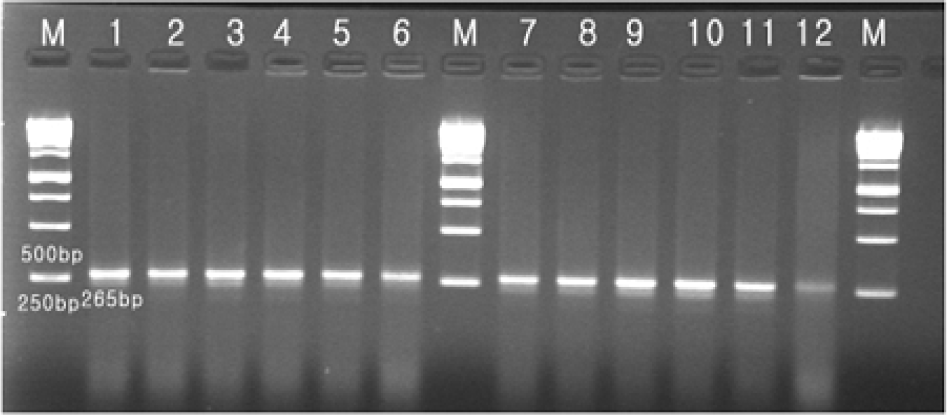 | Figure 2.Agarose gel electrophoresis of PCR-generated amplicons by using primer pairs for gyrA gene for detection of ciprofloxacin-resistant C. coli. Lane M: size marker, lane 1 to 12: isolate no. C01-1 to C01-12. |
Table 1.
Primers used to identify Campylobacter coli and to detect mutations in gyrA and nucleotide 2074 in 23S rRNA
Table 2.
Prevalence of Campylobacter coli isolates and their mutation rate in 23S rRNA A2074C
| Te st | No. of specimen tested | No. (%) positive |
|---|---|---|
| C. coli isolation | 100 | 55 (55) |
| 23S rRNA A2074C mutation detection | 55 | 23 (41.8) |
Table 3.
Phenotypic and genetic characteristics of Campylobacter coli isolate
| Isolate no./source | Hippurate hydrolysis | MIC (μg/ml) of: | Detection by PCR and mutation | Erythromycin MIC (μg/ml) | ||||
|---|---|---|---|---|---|---|---|---|
| Nalidixic acid | Ciprofloxacin | hipO | glyA | gyrA mutation | A2074C mutationa | |||
| C01-1 | – | 128 | 16 | – | + | + | – | 4 |
| C01-2 | – | 128 | 16 | – | + | + | – | 8 |
| C01-3 | – | 64 | 32 | – | + | + | + | ≥128 |
| C01-4 | – | 128 | 4 | – | + | + | – | 4 |
| C01-5 | – | 32 | 8 | – | + | + | + | ≥128 |
| C01-6 | – | 128 | 64 | – | + | + | – | 4 |
| C01-7 | – | 64 | 64 | – | + | + | + | ≥128 |
| C01-8 | – | 128 | 8 | – | + | + | – | 1 |
| C01-9 | – | 128 | 8 | – | + | + | + | ≥128 |
| C01-10 | – | 32 | 4 | – | + | + | – | ≤0.5 |
| C01-11 | – | 32 | 32 | – | + | + | + | ≥128 |
| C01-12 | – | 64 | 32 | – | + | + | + | ≥128 |
| C01-13 | – | 128 | 32 | – | + | + | – | 4 |
| C01-14 | – | 64 | 32 | – | + | + | – | ≤0.5 |
| C01-15 | – | 64 | 32 | – | + | + | – | ≤0.5 |
| C01-16 | – | 64 | 16 | – | + | + | – | ≤0.5 |
| C01-17 | – | 64 | 32 | – | + | + | + | ≥128 |
| C01-18 | – | 32 | 8 | – | + | + | – | 2 |
| C01-19 | – | 64 | 8 | – | + | + | – | 4 |
| C01-20 | – | 64 | 16 | – | + | + | – | 4 |
| C01-21 | – | 64 | 4 | – | + | + | + | ≥128 |
| C01-22 | – | 64 | 32 | – | + | + | + | ≥128 |
| C01-23 | – | 128 | 32 | – | + | + | – | 4 |
| C01-24 | – | 64 | 8 | – | + | + | – | 4 |
| C01-25 | – | 64 | 8 | – | + | + | – | 4 |
| C01-26 | – | 64 | 32 | – | + | + | + | ≥128 |
| C01-27 | – | 32 | 8 | – | + | + | + | ≥128 |
| C01-28 | – | 64 | 32 | – | + | + | + | ≥128 |
| C01-29 | – | 64 | 16 | – | + | + | + | ≥128 |
| C01-30 | – | 128 | 32 | – | + | + | – | 8 |
| C01-31 | – | 128 | 32 | – | + | + | – | 4 |
| C01-32 | – | 128 | 16 | – | + | + | – | 4 |
| C01-33 | – | 128 | 16 | – | + | + | – | 4 |
| C01-34 | – | 64 | 32 | – | + | + | + | ≥128 |
| C01-35 | – | 128 | 16 | – | + | + | + | ≥128 |
| C01-36 | – | 64 | 16 | – | + | + | + | ≥128 |
| C01-37 | – | 64 | 16 | – | + | + | + | ≥128 |
| C01-38 | – | 128 | 16 | – | + | + | – | 16 |
| C01-39 | – | 64 | 16 | – | + | + | + | ≥128 |
| C01-40 | – | 128 | 16 | – | + | + | – | 4 |
| C01-41 | – | 128 | 32 | – | + | + | – | 2 |
| C01-42 | – | 32 | 16 | – | + | + | – | ≤0.5 |
| C01-43 | – | 128 | 16 | – | + | + | – | 1 |
| C01-44 | – | 64 | 4 | – | + | + | – | 2 |
| C01-45 | – | 32 | 32 | – | + | + | + | 128 |
| C01-46 | – | 32 | 4 | – | + | + | – | 2 |
| C01-47 | – | 64 | 32 | – | + | + | + | ≥128 |
| C01-48 | – | 32 | 4 | – | + | + | – | ≤0.5 |
| C01-49 | – | 128 | 16 | – | + | + | – | 8 |
| C01-50 | – | 128 | 32 | – | + | + | – | 8 |
| C01-51 | – | 64 | 16 | – | + | + | + | ≥128 |
| C01-52 | – | 128 | 16 | – | + | + | – | 4 |
| C01-53 | – | 128 | 32 | – | + | + | + | ≥128 |
| C01-54 | – | 64 | 32 | – | + | + | – | 2 |
| C01-55 | – | 128 | 64 | – | + | + | + | ≥128 |
Table 4.
Comparision of antimicrobial susceptibilities of Campylobacter coli isolates with and without 23S rRNA A2074 mutation
| Antimicrobial agents | Isolate with 23S rRNA A2074C mutation | |||||
|---|---|---|---|---|---|---|
| Positive (n = 23) | Negative (n = 32) | |||||
| Sa | I | R | S | I | R | |
| Cefotaxime | 78 | 22 | 0 | 78 | 19 | 3 |
| Chloramphenicol | 96 | 0 | 4 | 97 | 0 | 3 |
| Amikacin | 56 | 0 | 44 | 75 | 9 | 16 |
| Gentamicin | 48 | 0 | 52 | 100 | 0 | 0 |
| Kanamycin | 9 | 0 | 91 | 9 | 16 | 75 |
| Cotrimoxazole | 4 | 0 | 96 | 13 | 0 | 87 |
Table 5.
MIC of antimicrobial agents for Campylobacter coli isolates with and without 23S rRNA A2074C mutation




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download