Abstract
The mouse model is alleged to be a useful tool for understanding of pathophysiological roles of Helicobacter pylori in the development of gastric disorders. However, it has been observed that H. pylori strains significantly differed in their fitness in mice and even mouse strains differed in their susceptibilities to a H. pylori strain. Bacterial components of H. pylori which could affect on its fitness in mice have to be elucidated for the establishment of the mouse model for H. pylori infections. In the comparison of colonization ability between two H. pylori Korean isolates, 51 (isolated from a patient with duodenal ulcer) and 52 (isolated from a patient with gastric cancer), 52 could colonize better than 51 on the gastric mucosa of mouse. Proteome components of H. pylori 52, as a good colonizer and H. pylori 51, as a poor one were quantitatively compared each other. Five bacterial proteins including catalase, urease subunit alpha/beta, enolase and ferritin, were up-regulated in 52. In addition, the respective proteome components of the two strains were also compared with their mouse-passaged homologous strains. Seven and five proteins, which included catalase, flagellin A/B in common, were up-regulated in mouse-adapted 51 and 52, respectively. Among the fourteen identified proteins, urease subunit alpha/beta, flagellin A/B, catalase, ferritin, superoxide dismutase and neutrophil-activation protein have been previously known to be necessary to gastric colonization of H. pylori in animal models. The other up-regulated proteins including enolase, elongation factor Tu and fructose-bisphosphate aldolase have been reported to be associated with acid tolerance of H. pylori. These data provide confirmatory evidence for the importance of those proteins in the development of H. pylori-associated gastric disorders.
Go to : 
REFERENCES
1). Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984; 1:1311–5.

3). Marshall BJ, Armstrong JA, McGechie DB, Glancy RJ. Attempt to fulfill Koch's postulates for pyloric Campylobacter. Med J Aust. 1985; 142:436–9.
4). World Health Organization. Schistosomes, liver flukes and Helicobacter pylori. IARC Monographs on the Evaluation of Calcinogenic Risks to Human. 1994. 177–240.
5). Baik SC, Youn HS, Chung MH, Lee WK, Cho MJ, Ko GH, et al. Increased oxidative DNA damage in Helicobacter pylori-infected human gastric mucosa. Cancer Res. 1996; 56:1279–82.
6). Baik SC, Kim JB, Cho MJ, Kim YC, Park CK, Ryou HH, et al. Prevalence of Helicobacter pylori infection among normal Korean adults. J Korean Soc Microbiol. 1990; 25:455–62.
7). Rhee KH, Youn HS, Baik SC, Lee WK, Cho MJ, Choi HJ, et al. Prevalence of Helicobacter pylori infection in Korea. J Korean Soc Microbiol. 1990; 25:475–90.
8). Youn HS, Baik SC, Lee WK, Cho MJ, Ryou HH, Choi HJ, et al. Serodiagnosis of Helicobacter pylori infection. J Korean Soc Microbiol. 1990; 25:463–74.
9). Youn HS, Ko GH, Chung MH, Lee WK, Cho MJ, Rhee KH. Pathogenesis and prevention of stomach cancer. J Korean Med Sci. 1996; 11:373–85.

10). Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997; 388:539–47.
11). Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999; 397:176–80.
12). Kawai M, Furuta Y, Yahara K, Tsuru T, Oshima K, Handa N, et al. Evolution in an oncogenic bacterial species with extreme genome plasticity: Helicobacter pylori East Asian genomes. BMC Microbiol. 2011; 11:104.

13). Oh JD, Kling-Bäckhed H, Giannakis M, Xu J, Fulton RS, Fulton LA, et al. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc Natl Acad Sci U S A. 2006; 103:9999–10004.
14). Baik SC, Kim KM, Song SM, Kim DS, Jun JS, Lee SG, et al. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of Helicobacter pylori strain 26695. J Bacteriol. 2004; 186:949–55. Erratum in: J Bacteriol 2005;187: 1541.
15). Bumann D, Aksu S, Wendland M, Janek K, Zimny-Arndt U, Sabarth N, et al. Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori. Infect Immun. 2002; 70:3396–403.
16). Cho MJ, Jeon BS, Park JW, Jung TS, Song JY, Lee WK, et al. Identifying the major proteome components of Helicobacter pylori strain 26695. Electrophoresis. 2002; 23:1161–73.
17). Park JW, Song JY, Lee SG, Jun JS, Park JU, Chung MJ, et al. Quantitative analysis of representative proteome components and clustering of Helicobacter pylori clinical strains. Helicobacter. 2006; 11:533–43.
18). Park JW, Lee SG, Song JY, Joo JS, Chung MJ, Kim SC, et al. Proteomic analysis of Helicobacter pylori cellular proteins fractionated by ammonium sulfate precipitation. Electrophoresis. 2008; 29:2891–903.
19). Krakowka S, Morgan DR, Kraft WG, Leunk RD. Establishment of gastric Campylobacter pylori infection in the neonatal gnotobiotic piglet. Infect Immun. 1987; 55:2789–96.
20). Radin MJ, Eaton KA, Krakowka S, Morgan DR, Lee A, Otto G, et al. Helicobacter pylori gastric infection in gnotobiotic beagle dogs. Infect Immun. 1990; 58:2606–12.
21). Karita M, Kouchiyama T, Okita K, Nakazawa T. New small animal model for human gastric Helicobacter pylori infection: success in both nude and euthymic mice. Am J Gastroenterol. 1991; 86:1596–603.
22). Marchetti M, Aricò B, Burroni D, Figura N, Rappuoli R, Ghiara P. Development of a mouse model of Helicobacter pylori infection that mimics human disease. Science. 1995; 267:1655–8.
23). Lee A, O'Rourke J, De Ungria MC, Robertson B, Daskalopoulos G, Dixon MF. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997; 112:1386–97. Erratum in: Gastroenterology 1997;113: 732.
24). Matsumoto S, Washizuka Y, Matsumoto Y, Tawara S, Ikeda F, Yokota Y, et al. Induction of ulceration and severe gastritis in Mongolian gerbil by Helicobacter pylori infection. J Med Microbiol. 1997; 46:391–7.
25). van Doorn NE, Namavar F, Sparrius M, Stoof J, van Rees EP, van Doorn LJ, et al. Helicobacter pylori-associated gastritis in mice is host and strain specific. Infect Immun. 1999; 67:3040–6.
26). Akada JK, Ogura K, Dailidiene D, Dailide G, Cheverud JM, Berg DE. Helicobacter pylori tissue tropism: mouse-colonizing strains can target different gastric niches. Microbiology. 2003; 149:1901–9.
27). Heukeshoven J, Dernick R. Improved silver staining procedure for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis. 1988; 9:28–32.

28). O'Connell KL, Stults JT. Identification of mouse liver proteins on two-dimensional electrophoresis gels by matrix-assisted laser desorption/ionization mass spectrometry of in situ enzymatic digests. Electrophoresis. 1997; 18:349–59.
29). Kim YH, Han KY, Lee K, Heo JH, Kang HA, Lee J. Comparative proteome analysis of Hansenula polymorpha DL1 and A16. Proteomics. 2004; 4:2005–13.
30). Shaw AC, Gevaert K, Demol H, Hoorelbeke B, Vandekerckhove J, Larsen MR, et al. Comparative proteome analysis of Chlamydia trachomatis serovar A, D and L2. Proteomics. 2002; 2:164–86.
31). Hunt SM, Thomas MR, Sebastian LT, Pedersen SK, Harcourt RL, Sloane AJ, et al. Optimal replication and the importance of experimental design for gel-based quantitative proteomics. J Proteome Res. 2005; 4:809–19.

32). Harris AG, Wilson JE, Danon SJ, Dixon MF, Donegan K, Hazell SL. Catalase (KatA) and KatA-associated protein (KapA) are essential to persistent colonization in the Helicobacter pylori SS1 mouse model. Microbiology. 2003; 149:665–72.
33). Harris AG, Hinds FE, Beckhouse AG, Kolesnikow T, Hazell SL. Resistance to hydrogen peroxide in Helicobacter pylori: role of catalase (KatA) and Fur, and functional analysis of a novel gene product designated ‘KatA-associated protein’, KapA (HP0874). Microbiology. 2002; 148:3813–25.
34). Seyler RW Jr, Olson JW, Maier RJ. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect Immun. 2001; 69:4034–40.
35). Hu LT, Foxall PA, Russell R, Mobley HL. Purification of recombinant Helicobacter pylori urease apoenzyme encoded by ureA and ureB. Infect Immun. 1992; 60:2657–66.
36). Krishnamurthy P, Parlow M, Zitzer JB, Vakil NB, Mobley HL, Levy M, et al. Helicobacter pylori containing only cytoplasmic urease is susceptible to acid. Infect Immun. 1998; 66:5060–6.
37). Bauerfeind P, Garner R, Dunn BE, Mobley HL. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut. 1997; 40:25–30.
38). Tsuda M, Karita M, Morshed MG, Okita K, Nakazawa T. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect Immun. 1994; 62:3586–9.
39). Hazell SL, Lee A, Brady L, Hennessy W. Campylobacter pyloridis and gastritis: association with intercellular spaces and adaptation to an environment of mucus as important factors in colonization of the gastric epithelium. J Infect Dis. 1986; 153:658–63.
40). Eaton KA, Suerbaum S, Josenhans C, Krakowka S. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect Immun. 1996; 64:2445–8.
41). Ottemann KM, Lowenthal AC. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect Immun. 2002; 70:1984–90.
42). Waidner B, Greiner S, Odenbreit S, Kavermann H, Velayudhan J, Stähler F, et al. Essential role of ferritin Pfr in Helicobacter pylori iron metabolism and gastric colonization. Infect Immun. 2002; 70:3923–9.
43). Rieder G, Einsiedl W, Hatz RA, Stolte M, Enders GA, Walz A. Comparison of CXC chemokines ENA-78 and interleukin-8 expression in Helicobacter pylori-associated gastritis. Infect Immun. 2001; 69:81–8.
44). Thompson SA, Blaser MJ. Isolation of the Helicobacter pylori recA gene and involvement of the recA region in resistance to low pH. Infect Immun. 1995; 63:2185–93.
45). Dong Q, Hyde D, Herra C, Kean C, Murphy P, O'Morain CA, et al. Identification of genes regulated by prolonged acid exposure in Helicobacter pylori. FEMS Microbiol Lett. 2001; 196:245–9.
46). Slonczewski JL, McGee DJ, Phillips J, Kirkpatrick C, Mobley HL. pH-dependent protein profiles of Helicobacter pylori analyzed by two-dimensional gels. Helicobacter. 2000; 5:240–7.
47). Zhang YN, Ding SG, Huang LH, Zhang J, Shi YY, Zhong LJ. Comparative proteome analysis of Helicobacter pylori clinical strains by two-dimensional gel electrophoresis. J Zhejiang Univ Sci B. 2011; 12:820–7.
Go to : 
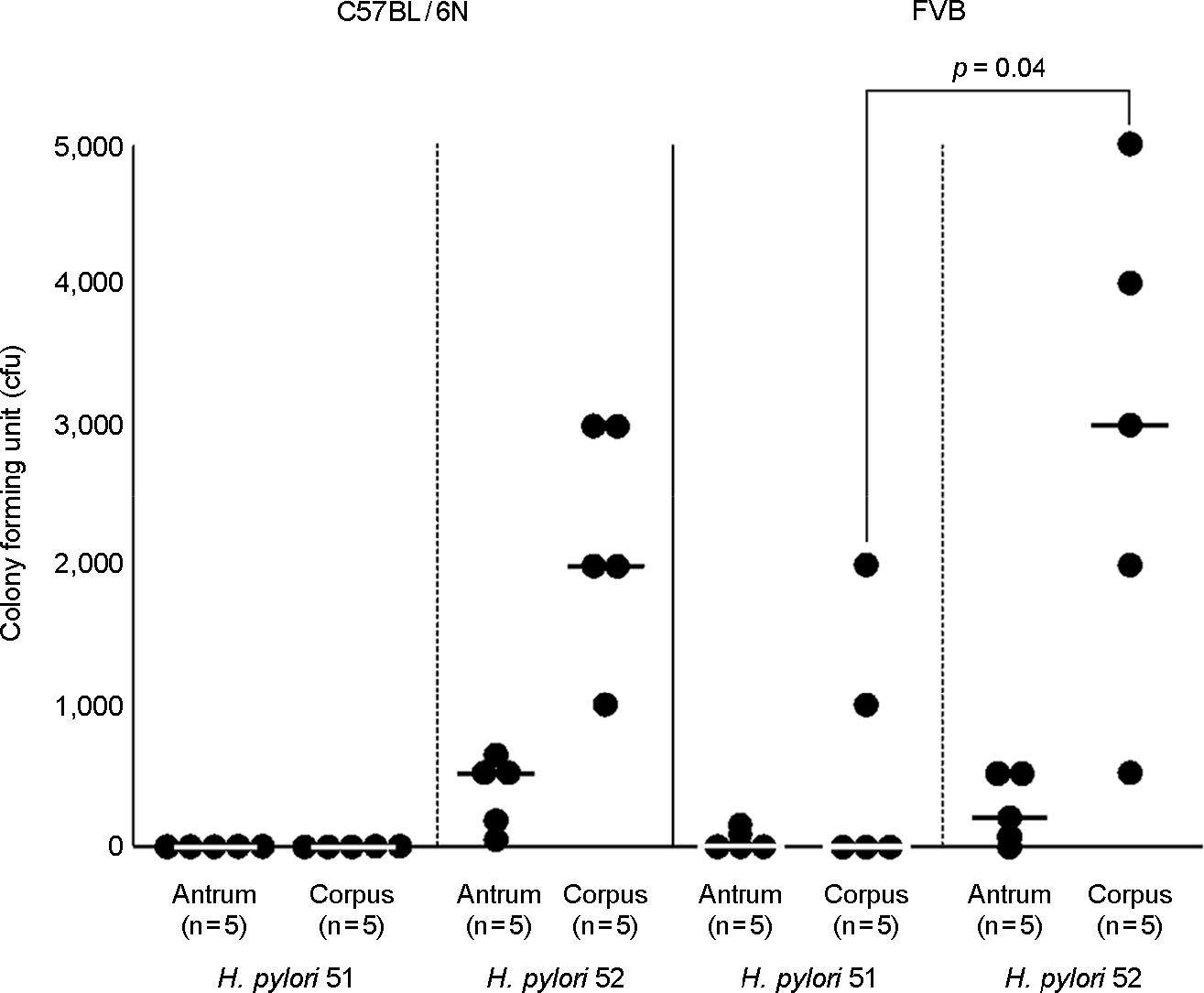 | Figure 1.Mouse colonization abilities of H. pylori 51 and 52. Mice were inoculated with H. pylori twice for two consecutive days with a dose of 2 × 109 cells. Mice were sacrificed 2 weeks later, antrum and corpus were separated and H. pylori was examined from each tissue by plate counts. Each spot represents the CFU count from one mouse. According to Mann-Whitney U test analysis, there is a significant difference between 51 and 52 in FVB corpus colonization ability (p = 0.04). The horizontal bars within each group represent the median value. |
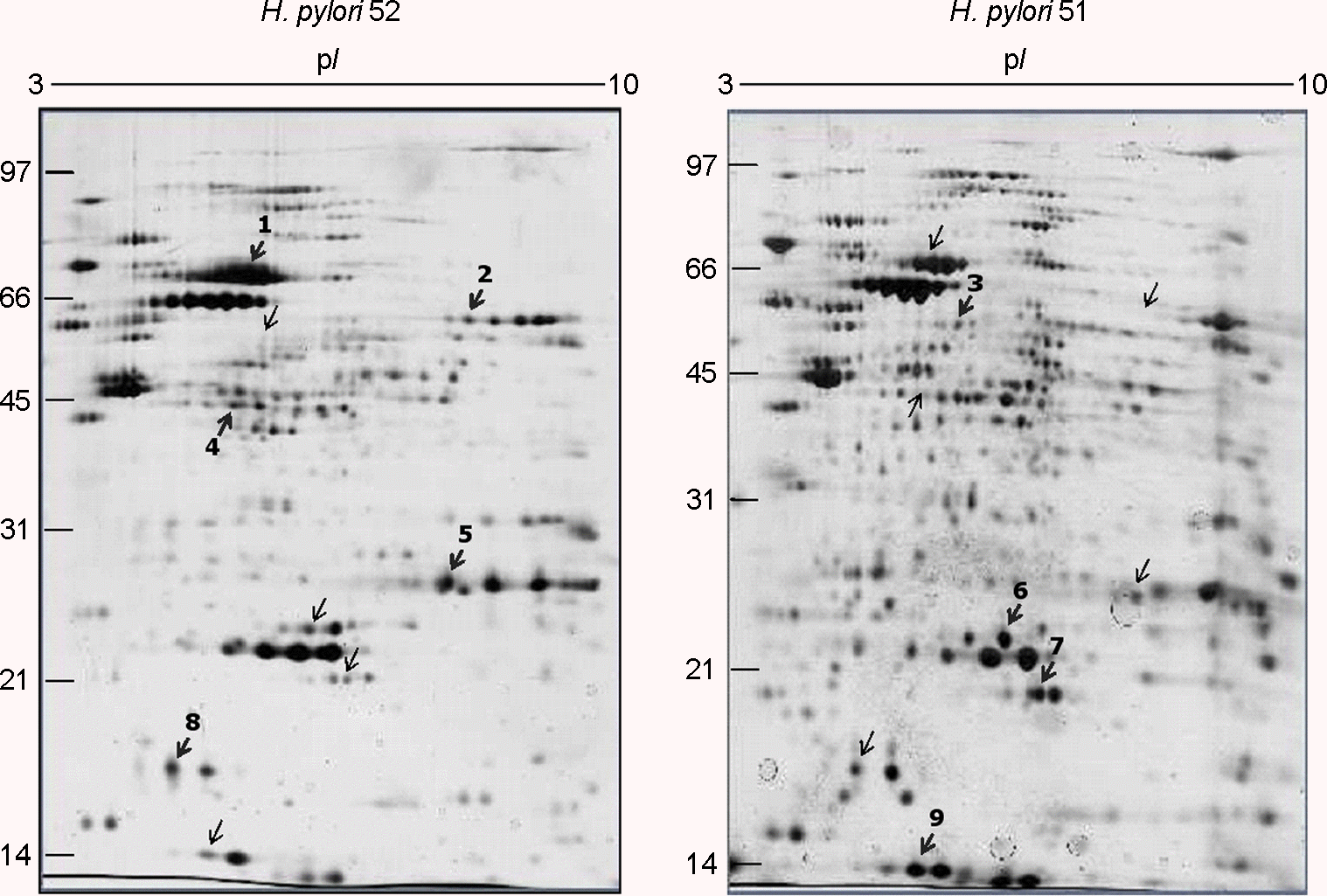 | Figure 2.Two-dimensional gel electrophoresis profiles of H. pylori 51 and 52. A total 100 μg of whole-cell protein of each strain was applied on 170-mm IPG strips with a range of pH 3–10, followed by 12% SDS-PAGE, and visualized by silver staining. The bold numbers with thick arrows represent proteins that are strong in spot intensity compared to the other strain. |
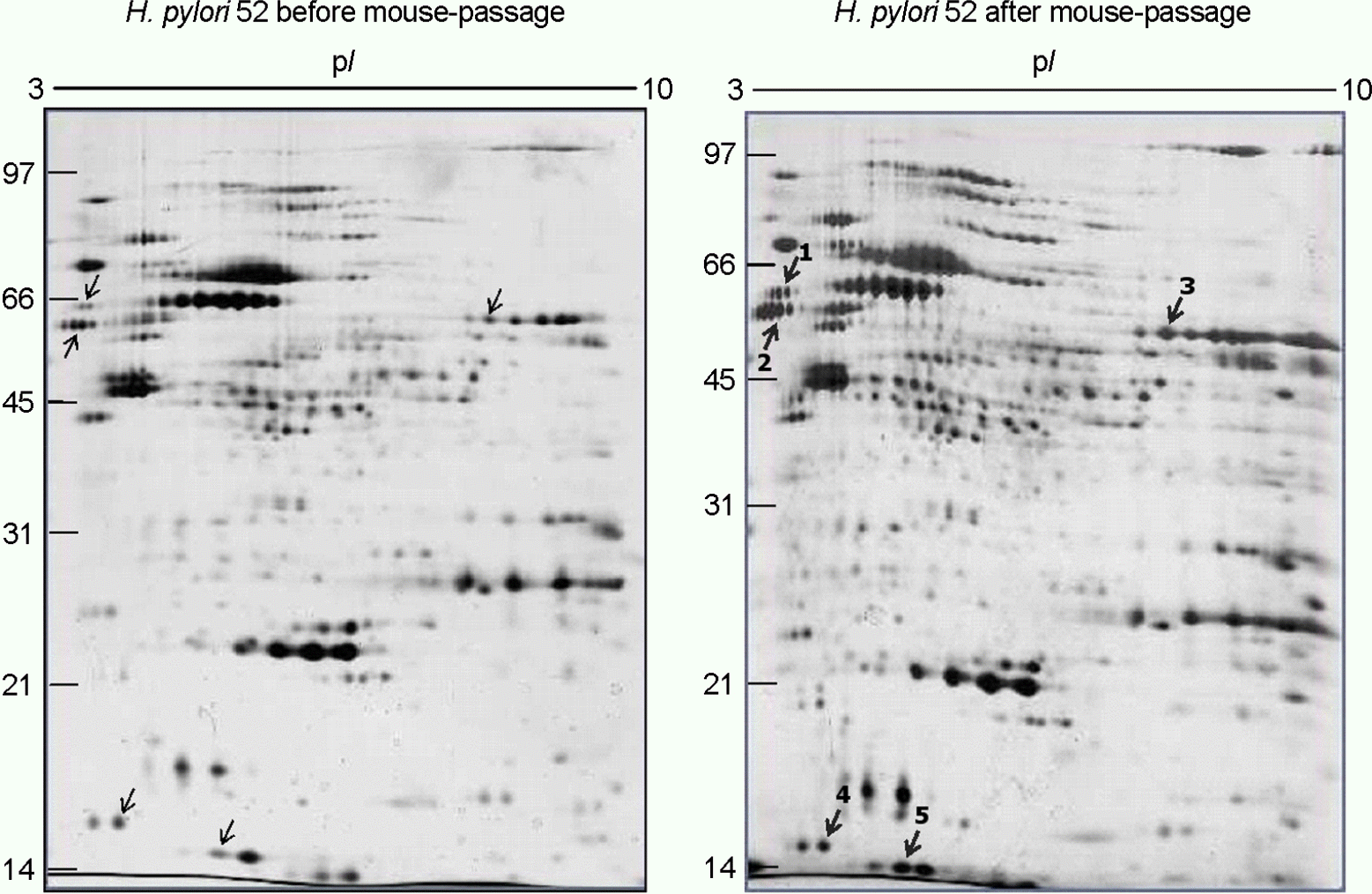 | Figure 3.Comparison of two-dimensional gel electrophoresis profiles of H. pylori 52 before and after FVB mouse passage. 100 μg of whole-cell protein of each strain was applied on 170-mm IPG strips with a range of pH 3∼10, followed by 12% SDS-PAGE, and visualized by silver staining. The bold numbers with thick arrows represent proteins that are up-regulated after mouse passage. |
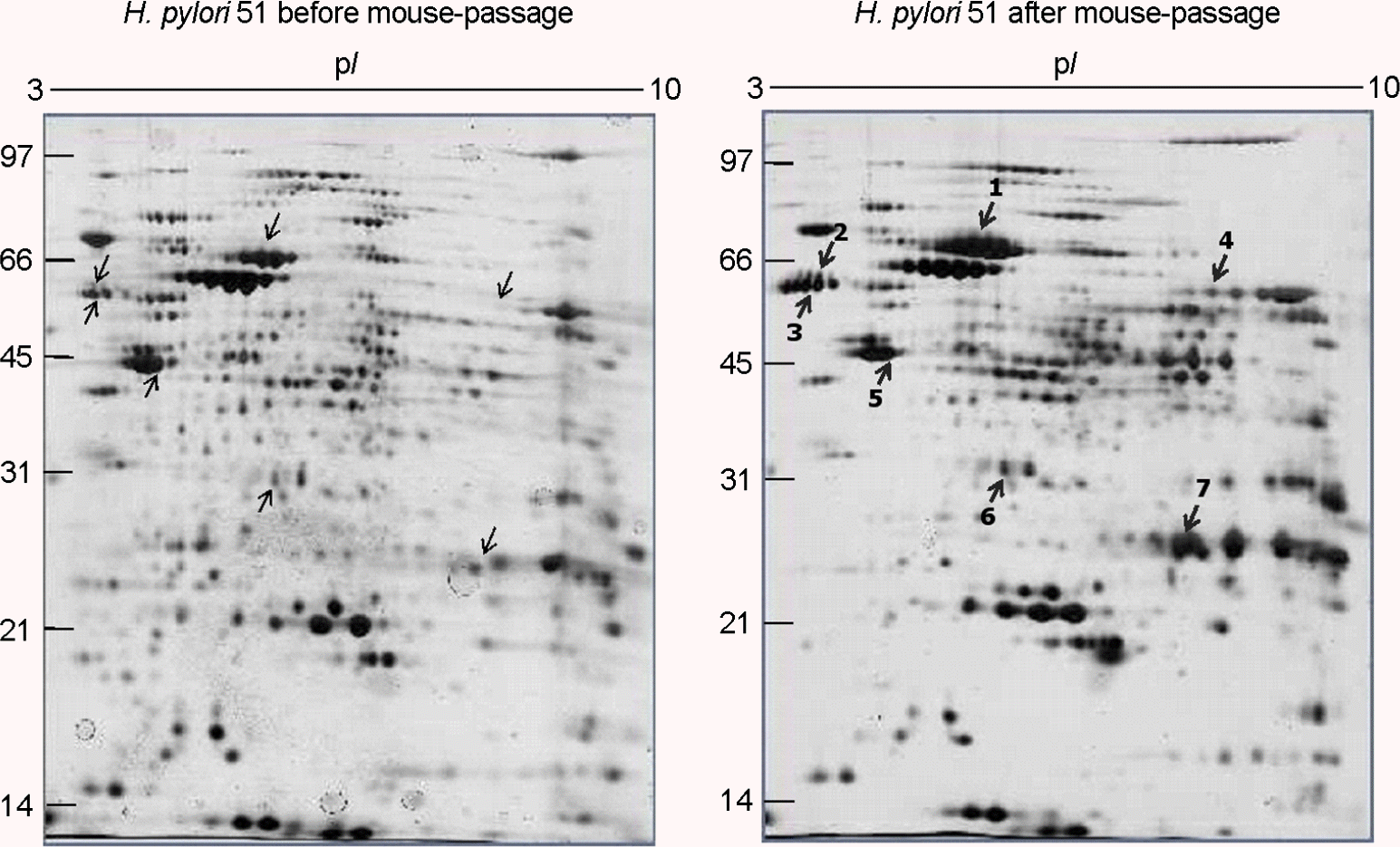 | Figure 4.Comparison of two-dimensional gel electrophoresis profiles of H. pylori 51 before and after FVB mouse passage. 100 μg of whole-cell protein of each strain was applied on 170-mm IPG strips with a range of pH 3∼10, followed by 12% SDS-PAGE, and visualized by silver staining. The bold numbers with thick arrows represent proteins that are up-regulated after mouse passage. |
Table 1.
Identification of differentially expressed proteins in the whole-cell protein fractions of H. pylori 51 and 52
| Spot No.* | Protein description | Gene codes | MW (Da) / pI |
Spot intensity (n=3, mean ± SD) |
Strain with up-regulation (fold variation) | |
|---|---|---|---|---|---|---|
| 51 | 52 | |||||
| 1 | Urease subunit beta | HP0072 | 61,683/5.9 | 498.40±376.97 | 1367.38±1013.2 | 52 (2.21) |
| 2 | Catalase | HP0875 | 58,630/8.7 | 57.14±59.41 | 185.74±51.35 | 52 (3.31) |
| 4 | Enolase | HP0154 | 46,562/5.5 | 18.63±32.26 | 180.33±18.31 | 52 (9.68) |
| 5 | Urease subunit alpha | HP0073 | 26,539/8.9 | 203.61±352.66 | 509.02±194.70 | 52 (2.50) |
| 8 | Ferritin | HP0653 | 1,9286/5.6 | 150.12±134.92 | 667.72±298.33 | 52 (4.45) |
| 3 | Glutamine synthetase | HP0512 | 54,513/6.0 | 67.55±28.73 | 3.91±6.78 | 51 (17.28) |
| 6 | Superoxide dismutase | HP0389 | 24,518/6.0 | 863.13±53.83 | 417.63±64.16 | 51 (2.07) |
| 7 | ATP-dependent protease binding subunit | HP0264 | 96,638/6.2 | 298.51±163.37 | 43.08±74.61 | 51 (6.93) |
| 9 | Neutrophil activating protein | HP0243 | 16,933/5.8 | 1404.04±309.76 | 324.69±283.82 | 51 (4.09) |
Table 2.
Identification of up-regulated proteins in the whole-cell protein fractions of H. pylori 52 after FVB mouse passage
Table 3.
Identification of up-regulated proteins in the whole-cell protein fractions of H. pylori 51 after FVB mouse passage




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download