Abstract
Gamma-glutamyltranspeptidase (GGT) was purified to electrophoretic homogeneity from the cell extract of H. pylori. The purified enzyme consisted of heavy and light subunits with molecular weights of 38 kDa and 21 kDa, respectively. N-terminal amino acid sequence of heavy and light subunits revealed that H. pylori GGT was processed into 3 parts for a signal peptide of 27 amino acid residues, a heavy subunit of 352 residues, and a light subunit of 188 residues during translation. The reaction rate for hydrolysis of γ-GpNA was 84.4 μmol/min per milligram of protein, and that for the γ-glutamyl transfer from γ-GpNA to gly-gly was 23.8 μmol/min per milligram of protein. The apparent Km values of H. pylori GGT for γ-glutamyl compounds were on the order of 10−3 to 10−4 M and those for acceptor peptides and amino acids were on the order of 10−1 to 10−2 M. The GGT protein kept approximately 80% of the initial enzymatic activity on incubation at 60°C for 15 min. The optimum temperature and pH for reactions of both hydrolysis and transpeptidation were 40°C and 9.0, respectively. The transpeptidation and hydrolysis reactions catalyzed by H. pylori GGT were strongly inhibited by L-Gln and moderately inhibited by L-Ala, L-Ser, β-chloro-L-Ala, and L-Glu. These results demonstrated that the biochemical properties of H. pylori GGT are different from those of other bacterial GGTs. Further, H. pylori GGT might degrade glutathione in the gastric mucous layer of humans if the enzyme could be secreted in the bacterial niches.
Go to : 
REFERENCES
1). Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984; 1:1311–5.

2). Blaser MJ. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J Infect Dis. 1990; 161:626–33.
3). Lee A, Fox J, Hazell S. Pathogenicity of Helicobacter pylori: a perspective. Infect Immun. 1993; 61:1601–10.
4). Correa P. Human gastric carcinogenesis: A multistep and multifactorial process-First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992; 52:6735–40.
5). Bagchi D, Bhattacharya G, Stohs SJ. Production of reactive oxygen species by gastric cells in association with Helicobacter pylori. Free Radic Res. 1996; 24:439–50.
6). Baik SC, Youn HS, Chung MH, Lee WK, Cho MJ, Ko GH, et al. Increased oxidative DNA damage in Helicobacter pylori-infected human gastric mucosa. Cancer Res. 1996; 56:1279–82.
7). Weitzman SA, Gordon LI. Inflammation and cancer: role of phagocyte-generated oxidants in carcinogenesis. Blood. 1990; 76:655–63.

8). Ghanayem BI, Boor PJ, Ahmed AE. Acrylonitrile-induced gastric mucosal necrosis: role of gastric glutathione. J Pharmacol Exp Ther. 1985; 232:570–7.
9). Giorgi G, Micheli L, Segre G, Pecchi A. Glutathione (GSH) in human stomach mucosa. Riv Eur Sci Med Farmacol. 1989; 11:163–7.
10). Orlowski M, Meister A. The gamma-glutamyl cycle: a possible transport system for amino acids. Proc Natl Acad Sci U S A. 1970; 67:1248–55.
11). Hiraishi H, Terano A, Ota S, Mutoh H, Sugimoto T, Harada T, et al. Protection of cultured rat gastric cells against oxidant-induced damage by exogenous glutathione. Gastroenterology. 1994; 106:1199–207.

12). Hirokawa K, Kawasaki H. Changes in glutathione in gastric mucosa of gastric ulcer patients. Res Commun Mol Pathol Pharmacol. 1995; 88:163–76.
13). Robert A, Eberle D, Kaplowitz N. Role of glutathione in gastric mucosal cytoprotection. Am J Physiol. 1984; 247:G296–304.

14). Shaw S, Herbert V, Colman N, Jayatilleke E. Effect of ethanol-generated free radicals on gastric intrinsic factor and glutathione. Alcohol. 1990; 7:153–7.

15). Shimizu M, Nakama A, Yamano T, Noda T, Fujita T, Kuroda K, et al. Role of gastric glutathione in smoke flavouring-induced gastric injury in rats. Food Chem Toxicol. 1992; 30:1005–9.

16). Szabo S, Nagy L, Plebani M. Glutathione, protein sulfhydryls and cysteine proteases in gastric mucosal injury and protection. Clin Chim Acta. 1992; 206:95–105.

17). Griffith OW, Meister A. Translocation of intracellular glutathione to membrane-bound gamma-glutamyl transpeptidase as a discrete step in the gamma-glutamyl cycle: glutathionuria after inhibition of transpeptidase. Proc Natl Acad Sci U S A. 1979; 76:268–72.
18). Griffith OW, Novogrodsky A, Meister A. Translocation of glutathione from lymphoid cells that have markedly different gamma-glutamyl transpeptidase activities. Proc Natl Acad Sci U S A. 1979; 76:2249–52.

19). Hanigan MH, Pitot HC. Gamma-glutamyl transpeptidaseits role in hepatocarcinogenesis. Carcinogenesis. 1985; 6:165–72.
20). Inoue M, Horiuchi S, Morino Y. gamma-Glutamyl transpeptidase in rat ascites tumor cells LY-5. Lack of functional correlation of its catalytic activity with the amino acid transport. Eur J Biochem. 1977; 78:609–15.
21). Rosalki SB, Rowe JA. Gamma-glutamyl-transpeptidase activity of human seminal fluid. Lancet. 1973; 1:323–4.

22). Tate SS, Meister A. gamma-Glutamyl transpeptidase: catalytic, structural and functional aspects. Mol Cell Biochem. 1981; 39:357–68.
23). Tate SS, Meister A. gamma-Glutamyl transpeptidase from kidney. Methods Enzymol. 1985; 113:400–19.
24). Ishiye M, Yamashita M, Niwa M. Molecular cloning of the gamma-glutamyltranspeptidase gene from a Pseudomonas strain. Biotechnol Prog. 1993; 9:323–31.
25). Mineyama R, Mikami K, Saito K. Partial purification and some properties of gamma-glutamyl peptidehydrolysing enzyme from Actinobacillus actinomy-cetemcomitans. Microbios. 1995; 82:7–19.
26). Orlowski M, Meister A. Isolation of gamma-glutamyl transpeptidase from hog kidney. J Biol Chem. 1965; 240:338–47.
27). Suzuki H, Kumagai H, Tochikura T. gamma-Glutamyltranspeptidase from Escherichia coli K-12: purification and properties. J Bacteriol. 1986; 168:1325–31.
28). Xu K, Strauch MA. Identification, sequence, and expression of the gene encoding gamma-glutamyltranspeptidase in Bacillus subtilis. J Bacteriol. 1996; 178:4319–22.
29). Nakayama R, Kumagai H, Tochikura T. Purification and properties of gamma-glutamyltranspeptidase from Proteus mirabilis. J Bacteriol. 1984; 160:341–6.
30). Suzuki H, Hashimoto W, Kumagai H. Escherichia coli K-12 can utilize an exogenous gamma-glutamyl peptide as an amino acid source, for which gamma-glutamyltranspeptidase is essential. J Bacteriol. 1993; 175:6038–40.
31). Megraud F, Bonnet F, Garnier M, Lamouliatte H. Characterization of “Campylorbacter pyloridis” by culture, enzymatic profile, and protein content. J Clin Microbiol. 1985; 22:1007–10.
32). Chevalier C, Thiberge JM, Ferrero RL, Labigne A. Essential role of Helicobacter pylori gamma-glutamyl-transpeptidase for the colonization of the gasric mucosa of mice. Mol Microbiol. 1999; 31:1359–72.
33). McGovern KJ, Blanchard TG, Gutierrez JA, Czinn SJ, Krakowka S, Youngman P. gamma-Glutamyltransferase is a Helicobacter pylori virulence factor but is not essential for colonization. Infect Immun. 2001; 69:4168–73.
34). Busiello I, Acquaviva R, Di Popolo A, Blanchard TG, Ricci V, Romano M, et al. Helicobacter pylori gamma-glutamyltranspeptidase upregulates COX-2 and EGF-related peptide expression in human gastric cells. Cell Microbiol. 2004; 6:255–67.
35). Kim KM, Lee SG, Park MG, Song JY, Kang HL, Lee WK, et al. Gamma-glutamyltranspeptidase of Helicobacter pylori induces mitochondria-mediated apoptosis in AGS cells. Biochem Biophys Res Commun. 2007; 355:562–7.
36). Kim KM, Lee SG, Kim JM, Kim DS, Song JY, Kang HL, et al. Helicobacter pylori gamma-glutamyl-transpeptidase induces cell cycle arrest at the G1-S phase transition. J Microbiol. 2010; 48:372–7.
37). Boanca G, Sand A, Barycki JJ. Uncoupling the enzymatic and autoprocessing activities of Helicobacter pylori gamma-glutamyltranspeptidase. J Biol Chem. 2006; 281:19029–37.
38). Boanca G, Sand A, Okada T, Suzuki H, Kumagai H, Fukuyama K, et al. Autoprocessing of Helicobacter pylori gamma-glutamyltranspeptidase leads to the formation of a threonine-threonine catalytic dyad. J Biol Chem. 2007; 282:534–41.
39). Shibayama K, Wachino J, Arakawa Y, Saidijam M, Rutherford NG, Henderson PJ. Metabolism of glutamine and glutathione via gamma-glutamyltranspeptidase and glutamate transport in Helicobacter pylori: possible significance in the pathophysiology of the organism. Mol Microbiol. 2007; 64:396–406.
40). Rhee KH, Lee WK, Baik SC, Cho MJ, Choi HJ. Production of the monoclonal antibody and the genomic library of Helicobacter pylori. J Korean Soc Microbiol. 1991; 26:305–16.
41). Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951; 193:265–75.

42). Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970; 227:680–5.

43). Moos M Jr, Nguyen NY, Liu TY. Reproducible high yield sequencing of proteins electrophoretically separated and transferred to an inert support. J Biol Chem. 1988; 263:6005–8.

45). Miller SP, Awasthi YC, Srivastava SK. Studies of human kidney gamma-glutamyl transpeptidase. Purification and structural, kinetic and immunological properties. J Biol Chem. 1976; 251:2271–8.

46). Suzuki H, Kumagai H, Echigo T, Tochikura T. DNA sequence of the Escherichia coli K-12 gamma-glutamyltranspeptidase gene, ggt. J Bacteriol. 1989; 171:5169–72.
47). Farinati F, Della Libera G, Cardin R, Molari A, Plebani M, Rugge M, et al. Gastric antioxidant, nitrites, and mucosal lipoperoxidation in chronic gastritis and Helicobacter pylori infection. J Clin Gastroenterol. 1996; 22:275–81.
Go to : 
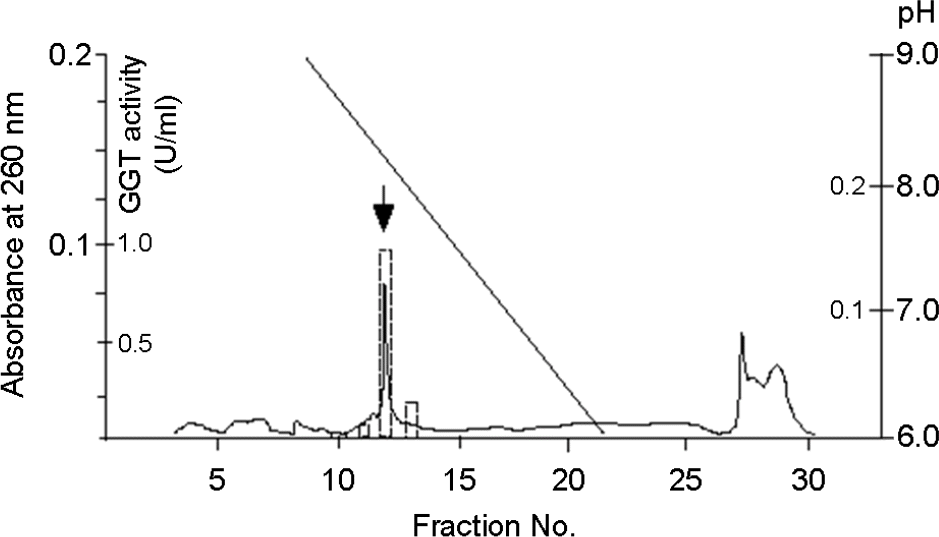 | Figure 1.Chromatofocusing pattern of H. pylori GGT. Enzyme fractions collected from a hydroxyapatite column were applied on a Mono P column (1 by 5 cm, Pharmacia) equilibrated with 25 mM ethanolamine-acetic acid buffer (pH 9.4) and eluted with 12 bed volumes of polybuffer 96-acetic acid (tenfold dilution [pH 6.0]) to form a descending linear gradient of pH 9 to 6 in the column. Dashed rectangle bars represented the enzyme activity (unit/m1) of fractions. The active fractions (arrow) eluted at pH 8.2 were pooled. |
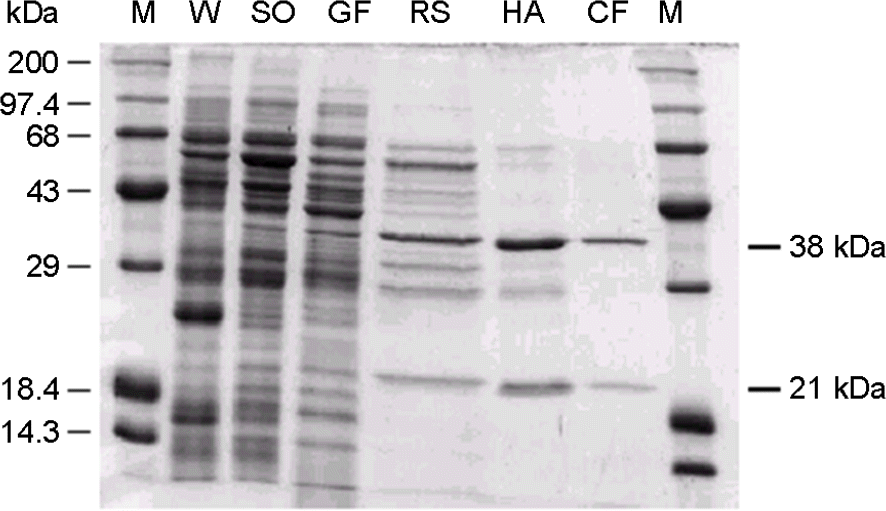 | Figure 2.SDS-polyacrylamide gel electrophoresis of H. pylori GGT in the purification steps. Approximately 8 μg of each fraction was applied. SDS-PAGE was performed with 12.5% polyacrylamide running gel (0.75 mm thick) and 3% polyacrylamide stacking gel at 15 mA for 2 hrs. After electrophoresis, a portion of the gel was stained with Coomassie brilliant blue R250. The subunit molecular weight was estimated by SDS-PAGE. Protein molecular weight standard, high range marker (200, 97.4, 68, 43, 29, 18.4, and 14.3 kDa) was used as a standard. M, marker; W, cell extract; SO, salt precipitation; GF, gel-filtration fraction; RS, Resource S fraction; HA, hydroxyapatite fraction; CF, chromatofocusing fraction. |
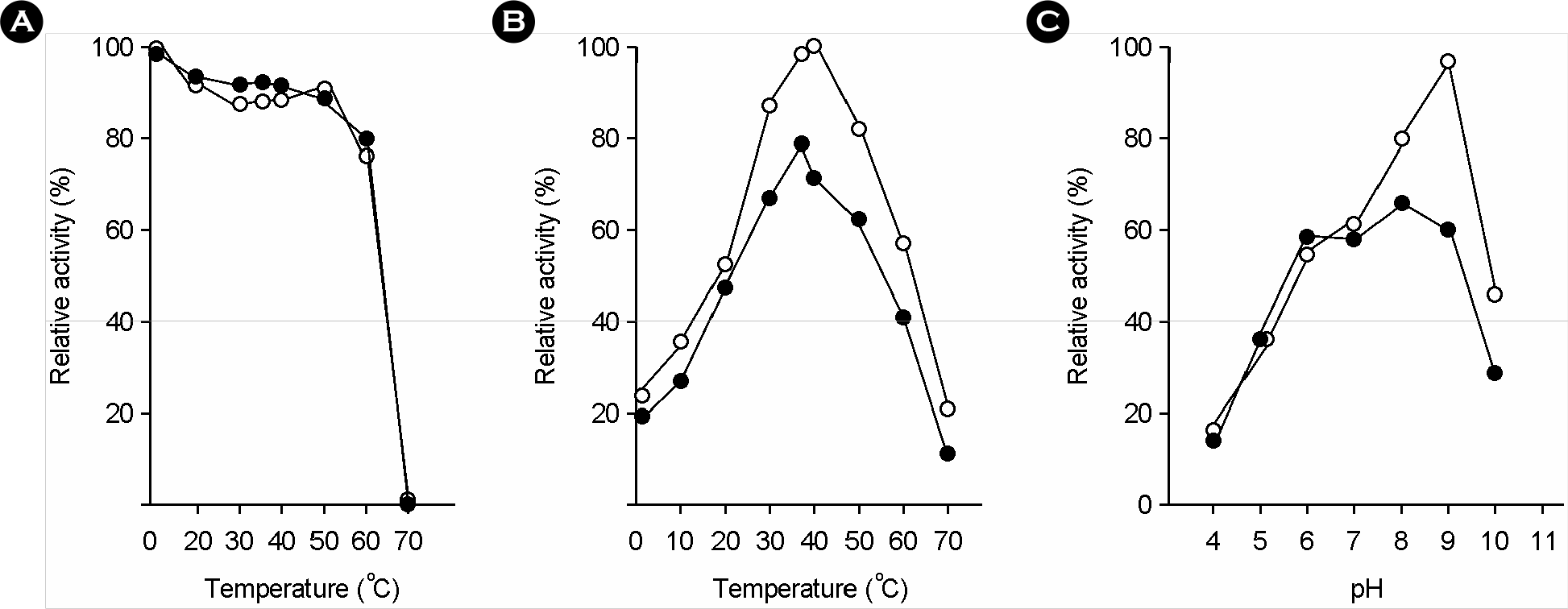 | Figure 3.Temperature stability and reactivity of H. pylori GGT at the various conditions of temperature and pH. (A) reaction mixtures were incubated at various temperatures for 15 min in 20 mM Tris-HCl (pH 8.0), and then GGT activities were measured; (B) GGT activities were measured at various temperature on the standard conditions described in Materials and Methods; (C) the pH of the reaction mixture containing the enzyme was adjusted with phosphate buffer for pH 4.0∼6.0 or Tris-HCl buffer for pH 7.0∼11.0 (open circles, transpeptidation activity; closed circles, hydrolysis activities). The indicated value is the mean of two independent experiments. |
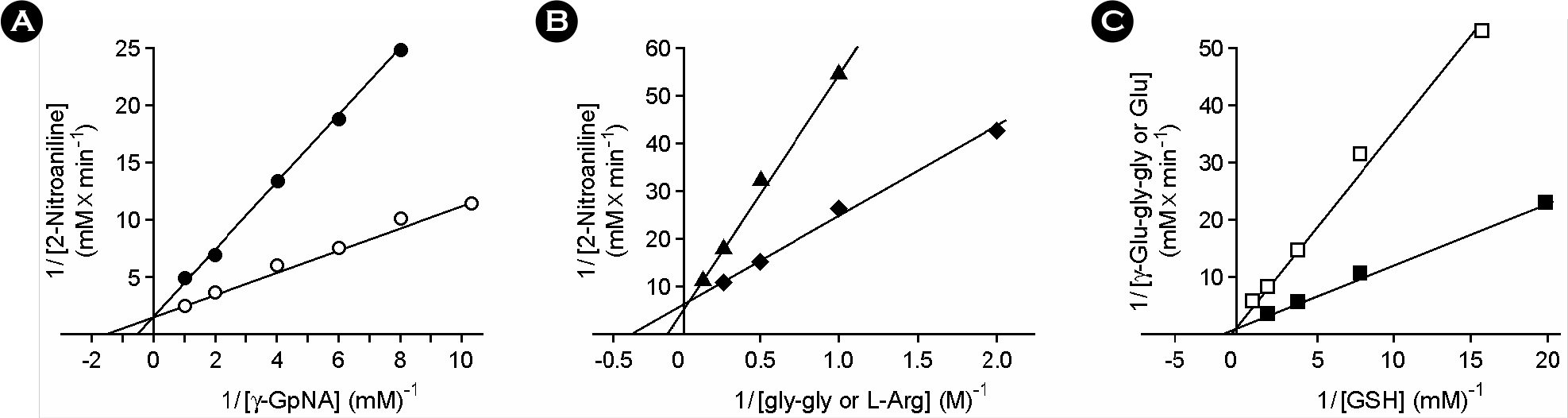 | Figure 4.Lineweaver-Burk plot of transpeptidation and/or hydrolysis of γ-GpNA, gly-gly, L-Arg, and GSH as catalyzed by purified H. pylori GGT. The enzyme activities were measured on the conditions described in Materials and Methods except for the change of the pH 9.0. (A) donor activities of γ-glutamyl moiety of γ-GpNA in transpeptidation reaction (○) and in hydrolysis reaction (•); (B) acceptor activities of gly-gly (⋄) and L-Arg (△) in transpeptidation reaction; C, donor activities of glutamyl moiety of GSH in transpeptidation reaction (□) and in hydrolysis reaction (▪). The reciprocals of the rates of 2-nitroaniline production (A, B) or γ-glutamyl-glycylglycine and glutamic acid production (C) were plotted against those of different concentrations of γ-glutamyl moiety donors or acceptors. The indicated values are the mean of two independent experiments. |
Table 1.
Summary of purification of GGT from H. pylori 51
Table 2.
Substrate specificity of H. pylori GGT for γ-glutamyl donors
| Substrates |
Relative activitya (%) |
|
|---|---|---|
| Transpeptidation | Hydrolysis | |
| γ-GpNA | 100b | 100 |
| GSH | 152.3 | 108.1 |
| GSSG | 229.3 | 127.8 |
| S-methyl GSH | 27.8 | 73.2 |
| γ-L-Glu-L-Tyr | 123.1 | 100.6 |
| γ-L-Glu-L-His | 147.9 | 110.3 |
| γ-L-Glu-L-Phe | 149.8 | 108.2 |
| γ-L-Glu-L-Leu | 147.7 | 136.8 |
| L-Gln | 137.8 | 110.1 |
| D-Gln | 389.7 | 105.5 |
| α-L-Glu-L-Ala | 0 | 0 |
Table 3.
Substrate specificity of H. pylori GGT for γ-glutamyl acceptors
| Substrates | Conc. (mM) |
Relative rate (%) |
||
|---|---|---|---|---|
| H. pylori | P. mirabilisa | E. colib | ||
| Gly-gly | 60 | 100 | 100 | 100 |
| Gly-gly | 20 | 62.4 | −c | 29.4 |
| L-Ala | 60 | 0 | 4.0 | 0 |
| L-α-Amino-n-butyrate | 60 | 0 | 5.5 | 12.7 |
| γ-Amino-n-butyrate | 60 | 54.0 | 16.3 | 22.3 |
| D-Arg | 60 | 37.2 | − | 18.1 |
| L-Arg | 60 | 79.9 | 20.7 | 141 |
| L-Asn | 60 | 38.7 | − | 31.8 |
| L-Asp | 60 | 0 | − | 4.2 |
| Benzoyl-L-Tyr ethyl ester | 20 | 0 | − | 0 |
| S-Carboxymethyl L-Cys | 20 | 0 | − | 1.9 |
| β-chloro-L-Alanine | 60 | 0 | 0 | − |
| D-Cys | 20 | 0 | − | 0 |
| L-Cys | 20 | 0 | 15.7 | 13.5 |
| L-Cysteic acid | 20 | 17.7 | − | 0 |
| L-DOPA | 20 | 0 | − | 63.9 |
| L-Gln | 60 | 0 | 2.5 | − |
| L-Glu | 60 | 0 | 0 | 10.2 |
| Gly | 60 | 0 | 16.6 | 20.8 |
| Gly-gly-gly | 60 | 29.7 | − | 14.2 |
| Gly-Leu | 60 | 0 | − | 12.5 |
| D-His | 60 | 0 | − | − |
| L-His | 60 | 0 | 36.9 | 49.5 |
| L-Hse | 60 | 0 | 6.7 | 0 |
| L-Ile | 60 | 19.8 | 25.2 | 13.8 |
| L-Leu | 20 | 17.1 | 20.7 | 2.5 |
| Leu-Gly | 60 | 0 | − | 12.5 |
| D-Lys | 60 | 55.0 | − | 16.5 |
| L-Lys | 60 | 46.6 | 16.0 | 121 |
| L-Met | 60 | 46.8 | 46.7 | 91.8 |
| 3-Methoxy-L-Tyr | 20 | 0 | − | − |
| S-Methyl-L-Cys | 20 | 0 | − | 13.5 |
| O-Methyl-L-Tyr | 20 | 0 | − | 5.6 |
| L-Phe | 60 | 52.6 | 51.7 | 42.9 |
| L-Pro | 60 | 50.0 | 13.2 | 2.4 |
| L-Ser | 60 | 0 | 6.2 | 9.6 |
| L-Thr | 60 | 0 | 15.6 | 22.9 |
| L-Trp | 20 | 30.7 | 36.9 | 29.8 |
| Trp-Gly | 20 | 0 | − | 23.6 |
| D-Tyr | 20 | 0 | − | 1.1 |
| L-Tyr | 20 | 20.4 | 6.6 | 2.8 |
| L-Val | 60 | 0 | 12.6 | 10.8 |
Table 4.
Km valuesa
| Parameters | Substrate | mM |
|---|---|---|
| Transferase activity | ||
| Donorb | γ-GpNA | 0.66±0.03 |
| GSH | 1.95±1.04 | |
| Acceptorc | Gly-gly | 32±6 |
| L-Arg | 107±6 | |
| Hydrolase activity | ||
| γ-GpNA | 1.64±0.06 | |
| GSH | 1.06±0.27 | |
Table 5.
Inhibition of H. pylori GGT by amino acids
| Amino acids |
Inhibition (%) of |
||
|---|---|---|---|
| Conc. (mM) | Transpeptidation | Hydrolysis | |
| None | 0a | 0 | |
| L-Ala | 10 | 56.1 | 54.8 |
| L-Ser | 10 | 48.3 | 48.4 |
| L-Hse | 10 | 36.3 | 18.9 |
| L-α-Ab | 10 | 16.1 | 20.9 |
| Gly | 10 | 22.9 | 25.8 |
| D-Cys | 10 | 72.1 | 12.4 |
| L-Cysteic acid | 10 | 72.8 | 3.9 |
| L-Asp | 10 | 22.3 | 12.0 |
| β-chloro-L-Ala | 10 | 57.0 | 42.3 |
| L-Cys | 10 | 61.3 | 11.2 |
| L-Gln | 10 | 91.7 | 89.8 |
| L-Glu | 10 | 72.7 | 46.5 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download