Abstract
In infants, urinary tract infections (UTIs) are quite common and primarily caused by bacterial pathogens. However, little research has been conducted regarding the relationship between uropathogenic bacteria, virulent genes, and uropathogenic viruses that might induce UTIs in infants. In this study, we evaluated infants with UTIs to determine the influence of bacterial virulent genes and type of viral infections on clinical aspects. First, we detected 44 cases of bacterial UTI from 600 suspected cases in infants and children. We detected E. coli urovirulence genes (kps, usp, pap, ireA, and cnf), two enteropathogenic E. coli genes (bfpA, and eae) and four S. aureus and S. epidermidis genes (mecA, pvl, bbp, and icaA) in urine samples from infant UTI cases. We also simultaneously detected hematuria-related adenovirus type 11, 21, and BK virus (BKV) in urine samples by PCR. As a result, E. coli was the most prevalent bacteria and in Dimercaptosuccinic acid (DMSA)-positive UTI cases, the uropathogenic E. coli virulence factor pap was significantly high. We found that BKV detection was significantly higher in DMSA-positive UTI infants (89%) compared with 50% of non-UTI (no bacteria detected) cases. These results are indicative of combined multiple bacterial and viral infections and show severe infant pyelonephritis.
REFERENCES
1). Hsiao AL, Chen L, Baker MD. Incidence and predictors of serious bacterial infections among 57- to 180-day-old infants. Pediatrics. 2006; 117:1695–701.

2). Chang SL, Shortliffe LD. Pediatric urinary tract infections. Pediatr Clin North Am. 2006; 53:379–400. vi.

4). Zorc JJ, Kiddoo DA, Shaw KN. Diagnosis and management of pediatric urinary tract infections. Clin Microbiol Rev. 2005; 18:417–22.

5). Marcus N, Ashkenazi S, Yaari A, Samra Z, Livni G. Non-Escherichia coli versus Escherichia coli community-acquired urinary tract infections in children hospitalized in a tertiary center: relative frequency, risk factors, antimicrobial resistance and outcome. Pediatr Infect Dis J. 2005; 24:581–5.
6). Prajapati BS, Prajapati RB, Patel PS. Advances in management of urinary tract infections. Indian J Pediatr. 2008; 75:809–14.

7). Johnson JR, Stapleton AE, Russo TA, Scheutz F, Brown JJ, Maslow JN. Characteristics and prevalence within serogroup O4 of a J96-like clonal group of uropathogenic Escherichia coli O4: H5 containing the class I and class III alleles of papG. Infect Immun. 1997; 65:2153–9.
8). Akiyama H, Kurosu T, Sakashita C, Inoue T, Mori Si, Ohashi K, et al. Adenovirus is a key pathogen in hemorrhagic cystitis associated with bone marrow transplantation. Clin Infect Dis. 2001; 32:1325–30.

9). Bedi A, Miller CB, Hanson JL, Goodman S, Ambinder RF, Charache P, et al. Association of BK virus with failure of prophylaxis against hemorrhagic cystitis following bone marrow transplantation. J Clin Oncol. 1995; 13:1103–9.

10). Azzi A, Cesaro S, Laszlo D, Zakrzewska K, Ciappi S, De Santis R, et al. Human polyomavirus BK (BKV) load and haemorrhagic cystitis in bone marrow transplantation patients. J Clin Virol. 1999; 14:79–86.

11). Park HK, Jung YJ, Chae HC, Shin YJ, Woo SY, Park HS, et al. Comparison of Escherichia coli uropathogenic genes (kps, usp and ireA) and enteroaggregative genes (aggR and aap) via multiplex polymerase chain reaction from suprapubic urine specimens of young children with fever. Scand J Urol Nephrol. 2009; 43:51–7.
12). Arisoy M, Aysev D, Ekim M, Ozel D, Köse SK, Ozsoy ED, et al. Detection of virulence factors of Escherichia coli from children by multiplex polymerase chain reaction. Int J Clin Pract. 2006; 60:170–3.
13). Kanamaru S, Kurazono H, Ishitoya S, Terai A, Habuchi T, Nakano M, et al. Distribution and genetic association of putative uropathogenic virulence factors iroN, iha, kpsMT, ompT and usp in Escherichia coli isolated from urinary tract infections in Japan. J Urol. 2003; 170:2490–3.
14). Russo TA, Carlino UB, Johnson JR. Identification of a new iron-regulated virulence gene, ireA, in an extra-intestinal pathogenic isolate of Escherichia coli. Infect Immun. 2001; 69:6209–16.
15). Persson S, Olsen KE, Scheutz F, Krogfelt KA, Gerner-Smidt P. A method for fast and simple detection of major diarrhoeagenic Escherichia coli in the routine diagnostic laboratory. Clin Microbiol Infect. 2007; 13:516–24.
16). Aranda KR, Fabbricotti SH, Fagundes-Neto U, Scaletsky IC. Single multiplex assay to identify simultaneously enteropathogenic, enteroaggregative, enterotoxigenic, enteroinvasive and Shiga toxin-producing Escherichia coli strains in Brazilian children. FEMS Microbiol Lett. 2007; 267:145–50.
17). Park HK, Woo SY, Jung YJ, Lee EO, Cha JE, Park HS, et al. Detection of virulence genes of Staphylococcus aureus and Staphylococcus epidermidis isolated from suprapubic urine from infants with fever. J Bacteriol Virol. 2008; 38:189–96.
18). Tristan A, Ying L, Bes M, Etienne J, Vandenesch F, Lina G. Use of multiplex PCR to identify Staphylococcus aureus adhesins involved in human hematogenous infections. J Clin Microbiol. 2003; 41:4465–7.
19). Arciola CR, Campoccia D, Gamberini S, Donati ME, Baldassarri L, Montanaro L. Occurrence of ica genes for slime synthesis in a collection of Staphylococcus epidermidis strains from orthopedic prosthesis infections. Acta Orthop Scand. 2003; 74:617–21.
20). Hatakeyama N, Suzuki N, Yamamoto M, Kuroiwa Y, Hori T, Mizue N, et al. Detection of BK virus and adenovirus in the urine from children after allogeneic stem cell transplantation. Pediatr Infect Dis J. 2006; 25:84–5.

21). Xu W, Erdman DD. Type-specific identification of human adenovirus 3, 7, and 21 by a multiplex PCR assay. J Med Virol. 2001; 64:537–42.

22). Trabulsi LR, Keller R, Tardelli Gomes TA. Typical and atypical enteropathogenic Escherichia coli. Emerg Infect Dis. 2002; 8:508–13.
23). Matar GM, Abdo D, Khneisser I, Youssef M, Zouheiry H, Abdelnour G, et al. The multiplex-PCR-based detection and genotyping of diarrhoeagenic Escherichia coli in diarrhoeal stools. Ann Trop Med Parasitol. 2002; 96:317–24.
24). Routh JC, Alt AL, Ashley RA, Kramer SA, Boyce TG. Increasing prevalence and associated risk factors for methicillin resistant Staphylococcus aureus bacteriuria. J Urol. 2009; 181:1694–8.
25). Decker DB, Karam JA, Wilcox DT. Pediatric hemorrhagic cystitis. J Pediatr Urol. 2009; 5:254–64.
26). Miyamura K, Takeyama K, Kojima S, Minami S, Matsuyama K, Morishima Y, et al. Hemorrhagic cystitis associated with urinary excretion of adenovirus type 11 following allogeneic bone marrow transplantation. Bone Marrow Transplant. 1989; 4:533–5.
27). Hatakeyama N, Suzuki N, Kudoh T, Hori T, Mizue N, Tsutsumi H. Successful cidofovir treatment of adenovirus-associated hemorrhagic cystitis and renal dysfunction after allogenic bone marrow transplant. Pediatr Infect Dis J. 2003; 22:928–9.
28). Fanourgiakis P, Georgala A, Vekemans M, Triffet A, De Bruyn JM, Duchateau V, et al. Intravesical instillation of cidofovir in the treatment of hemorrhagic cystitis caused by adenovirus type 11 in a bone marrow transplant recipient. Clin Infect Dis. 2005; 40:199–201.

30). Erdoğan O, Bülbül M, Demircin G, Oner A, Memis L. Acute necrotizing tubulointerstitial nephritis due to systemic adenoviral infection. Pediatr Nephrol. 2001; 16:265–8.
32). Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971; 1:1253–7.

33). Gonzalez JL, Balestra S, Schned AR, Gutmann EJ, Tsongalis GJ. Polyomavirus infection of the urinary tract presenting as hemorrhagic cystitis in an immunocompetent five-year-old boy. Diagn Cytopathol. 2008; 36:375–8.

34). Rocha SP, Elias WP, Cianciarullo AM, Menezes MA, Nara JM, Piazza RM, et al. Aggregative adherence of uropathogenic Proteus mirabilis to cultured epithelial cells. FEMS Immunol Med Microbiol. 2007; 51:319–26.
35). Coker C, Poore CA, Li X, Mobley HL. Pathogenesis of Proteus mirabilis urinary tract infection. Microbes Infect. 2000; 2:1497–505.
36). Rózalski A, Sidorczyk Z, Kotelko K. Potential virulence factors of Proteus bacilli. Microbiol Mol Biol Rev. 1997; 61:65–89.
Figure 1.
NuSieve agar gel showing detection of five urovirulent genes (kps, usp, pap, cnf, and ireA) and enteropathogenic genes of E. coli (eae and bfpA) in infant urine by multiplex PCR. Lane 1, DNA marker (50-bp ladder); Lane 2, kps (400 bp) and pap (328 bp) both positive; Lane 3, cnf (498 bp) and pap (328 bp) both positive; Lane 4, usp (440 bp) and pap (328 bp) both positive; Lane 5, usp (440 bp) and kps (400 bp) both positive; Lane 6, DNA marker (50-bp ladder); Lane 7, pap (328 bp) positive; Lane 8, usp (440 bp) and kps (400 bp) both positive; Lane 9, usp (440 bp) positive; Lane 10, kps (400 bp) positive; Lane 11, eae (377 bp) and bfpA (326 bp) both positive; Lane 12, DNA marker (50-bp ladder).
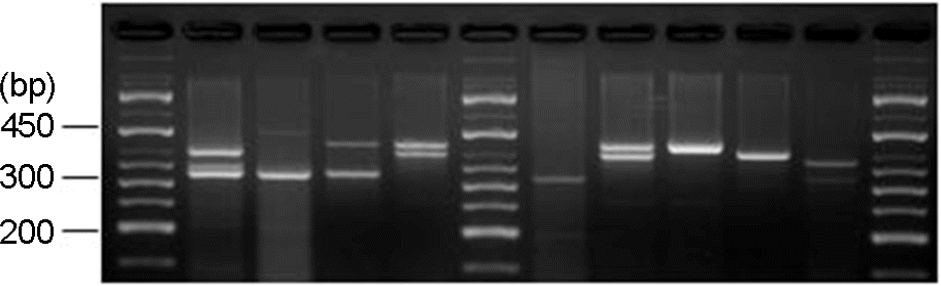
Figure 2.
NuSieve agar gel showing results of S. aureus and S. epidermidis virulence genes (mecA, pvl, bbp, and icaA) detected in infant urine by multiplex PCR. Lane 1, DNA marker (100-bp ladder); Lane 2, mecA (310 bp) positive; Lane 4, pvl (433 bp) positive; Lane 6, mecA (310 bp) positive; Lane 7, DNA marker (100-bp ladder); Lane 8, bbp (575 bp) and icaA (103 bp) both positive; Lane 12, DNA marker (100-bp ladder).
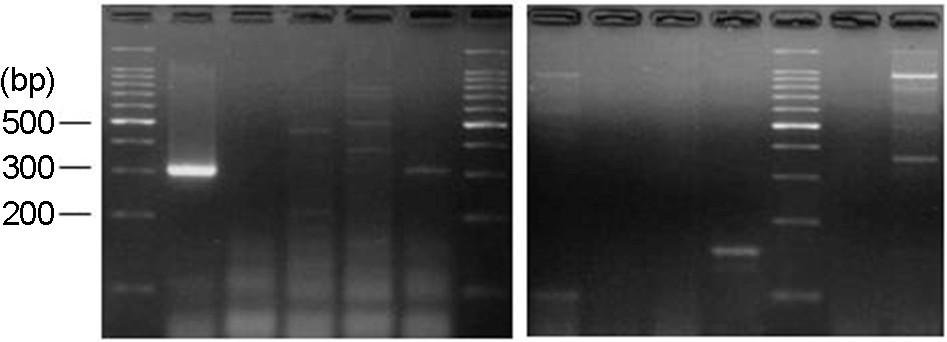
Figure 3.
NuSieve agar gel showing results of adenovirus type 11 (139 bp) detected in infant urine by monoplex PCR. Lane 1, DNA marker (123-bp ladder); Lane 4, AD11 positive; Lane 6, DNA marker (100-bp ladder); Lane 8, AD11 positive; Lane 11, AD11; Lane 12, DNA marker (100-bp ladder).
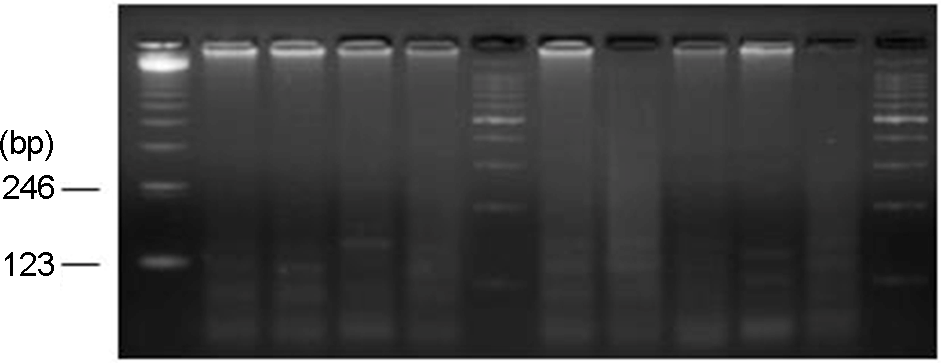
Figure 4.
NuSieve agar gel showing results of adenovirus type 21 (237 bp) detected in infant urine by monoplex PCR. Lane 1, DNA marker (100-bp ladder); Lane 3, AD21 positive; Lane 7, DNA marker (100-bp ladder); Lane 8, DNA marker (123-bp ladder); Lane 10, AD21 positive; Lane 11, AD21 positive; Lane 14, DNA marker (100-bp ladder).
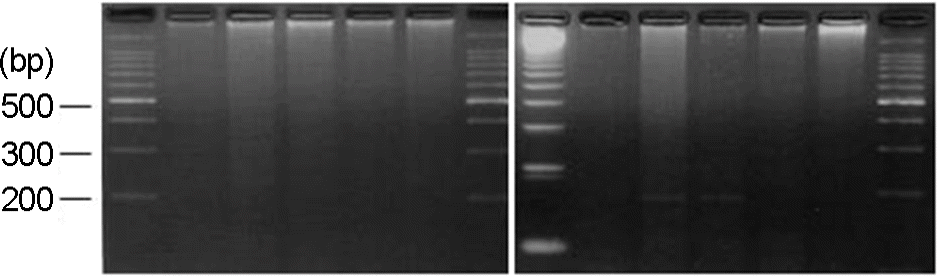
Figure 5.
NuSieve agar gel showing results of BKV (179 bp) detected in infant urine by monoplex PCR. Lane 1, DNA marker (100-bp ladder); Lane 2, BKV positive; Lane 3, BKV positive; Lane 4, BKV positive; Lane 5, BKV positive; Lane 6, DNA marker (100-bp ladder); Lane 7, BKV positive; Lane 8, BKV positive; Lane 9, BKV positive; Lane 11, BKV positive; Lane 12, DNA marker (100-bp ladder).
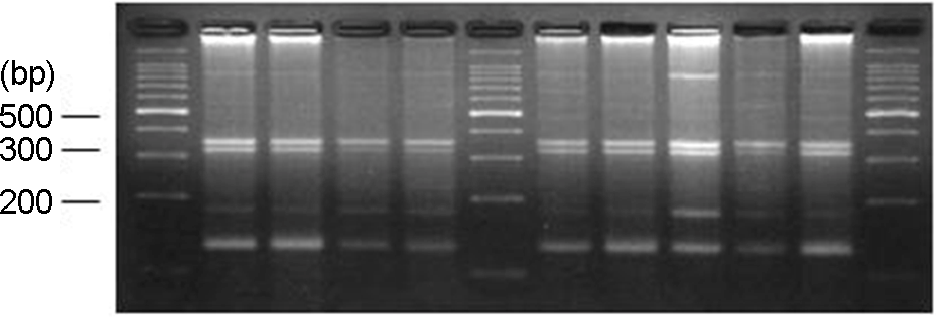
Table 1.
PCR primers for virulent genes used in this study
Table 2.
PCR primer sequences used in this study for adenovirus type 11, 21, and BKV
Table 3.
The relationship between DMSA-positive pyelonephritis and the detection rate of E. coli virulent genes
Table 4.
The relationship between DMSA-positive pyelonephritis and the detection rate of AD 11, A21 or BKV
Table 5.
The combined result of DMSA scan, the detection rate of virus, occult blood in the urine and E. coli virulent genes
Table 6.
The relationship between AD11, AD21, and BKV detection and occult blood in the urine




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download