Abstract
Inflammasome is a cytosolic multiprotein complex to activate caspase-1 leading to the subsequent processing of inactive pro-interleukin-1-beta (Pro-IL-1β) into its active interleukin-1 beta (IL-1β) in response to pathogen- or danger-associated molecular pattern. In recent years, a huge progress has been made to identify inflammasome component as a molecular platform to recruit and activate caspase-1. Nucleotide-binding oligomerization domain-like receptor (NLR) family proteins such as NLRP1, NLRP3 or interleukin-1β-converting enzyme (ICE)-protease activating factor (IPAF) have been first characterized to form inflammasome complex to induce caspase-1 activation. More recently, non-NLR type, pyrin-domain (PYD)-containing proteins such as pyrin or absent in melanoma2 (AIM2) were also proposed to form caspase-1-activating inflammasome machinery with apoptosis-associated speck-like protein containing a CARD (ASC), an essential adaptor molecule. Inflammasome pathways were shown to be crucial for protecting host organisms against diverse pathogen infections, but accumulating evidences also suggest that excessive activation of inflammasome/caspase-1 might be related to the pathogenesis of inflammation-related diseases. Indeed, mutations in NLRP3 or pyrin are closely associated with autoinflammatory diseases such as familial Mediterranean fever (FMF) syndrome or Muckle-Wells syndrome (MWS), indicating that the regulation of caspase-1 activity by inflammasome is a central process in these hereditary inflammatory disorders. Here, recent advances on the molecular mechanism of caspase-1 activation by PYD-containing inflammasomes are summarized and discussed.
Go to : 
REFERENCES
3). O'Neill LA., Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007. 7:353–64.
4). Trinchieri G., Sher A. Cooperation of Toll-like receptor signals in innate immune defence. Nat Rev Immunol. 2007. 7:179–90.

7). Seth RB., Sun L., Ea CK., Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005. 122:669–82.
8). Martinon F., Mayor A., Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009. 27:229–65.

9). Martinon F., Burns K., Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002. 10:417–26.
10). Yu JW., Fernandes-Alnemri T., Datta P., Wu J., Juliana C., Solorzano L, et al. Pyrin activates the ASC pyroptosome in response to engagement by autoinflammatory PSTPIP1 mutants. Mol Cell. 2007. 28:214–27.

11). Fernandes-Alnemri T., Yu JW., Datta P., Wu J., Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009. 458:509–13.

12). Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009. 458:514–8.

13). Bürckstümmer T., Baumann C., Blüml S., Dixit E., Dürnberger G., Jahn H, et al. An orthogonal proteomicgenomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009. 10:266–72.

14). Roberts TL., Idris A., Dunn JA., Kelly GM., Burnton CM., Hodgson S, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009. 323:1057–60.

15). Lamkanfi M., Declercq W., Kalai M., Saelens X., Vandenabeele P. Alice in caspase land. A phylogenetic analysis of caspases from worm to man. Cell Death Differ. 2002. 9:358–61.

16). Scott AM., Saleh M. The inflammatory caspases: guardians against infections and sepsis. Cell Death Differ. 2007. 14:23–31.

17). Li P., Allen H., Banerjee S., Franklin S., Herzog L., Johnston C, et al. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell. 1995. 80:401–11.
18). Lamkanfi M., Sarkar A., Vande Walle L., Vitari AC., Amer AO., Wewers MD, et al. Inflammasome-dependent release of the alarmin HMGB1 in endotoxemia. J Immunol. 2010. 185:4385–92.

19). Mariathasan S., Weiss DS., Dixit VM., Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med. 2005. 202:1043–9.
20). Miao EA., Leaf IA., Treuting PM., Mao DP., Dors M., Sarkar A, et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol. 2010. 11:1136–42.

22). Thornberry NA., Bull HG., Calaycay JR., Chapman KT., Howard AD., Kostura MJ, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992. 356:768–74.
23). Elliott JM., Rouge L., Wiesmann C., Scheer JM. Crystal structure of procaspase-1 zymogen domain reveals insight into inflammatory caspase autoactivation. J Biol Chem. 2009. 284:6546–53.

24). Srinivasula SM., Poyet JL., Razmara M., Datta P., Zhang Z., Alnemri ES. The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. J Biol Chem. 2002. 277:21119–22.

25). Poyet JL., Srinivasula SM., Tnani M., Razmara M., Fernandes-Alnemri T., Alnemri ES. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001. 276:28309–13.

26). Schotte P., Denecker G., Van Den Broeke A., Vandenabeele P., Cornelis GR., Beyaert R. Targeting Rac1 by the Yersinia effector protein YopE inhibits caspase-1-mediated maturation and release of interleukin-1beta. J Biol Chem. 2004. 279:25134–42.
27). Fernandes-Alnemri T., Wu J., Yu JW., Datta P., Miller B., Jankowski W, et al. The pyroptosome: a supra-molecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007. 14:1590–604.

28). Mariathasan S., Newton K., Monack DM., Vucic D., French DM., Lee WP, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004. 430:213–8.

29). Martinon F., Pétrilli V., Mayor A., Tardivel A., Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006. 440:237–41.

30). Mariathasan S., Weiss DS., Newton K., McBride J., O'Rourke K., Roose-Girma M, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006. 440:228–32.

31). Kanneganti TD., Ozöen N., Body-Malapel M., Amer A., Park JH., Franchi L, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006. 440:233–6.

32). Eisenbarth SC., Colegio OR., O'Connor W., Sutterwala FS., Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008. 453:1122–6.

33). Park HH., Lo YC., Lin SC., Wang L., Yang JK., Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007. 25:561–86.

34). Srimathi T., Robbins SL., Dubas RL., Chang H., Cheng H., Roder H, et al. Mapping of POP1-binding site on pyrin domain of ASC. J Biol Chem. 2008. 283:15390–8.

35). Hiller S., Kohl A., Fiorito F., Herrmann T., Wider G., Tschopp J, et al. NMR structure of the apoptosis- and inflammation-related NALP1 pyrin domain. Structure. 2003. 11:1199–205.

36). Moriya M., Taniguchi S., Wu P., Liepinsh E., Otting G., Sagara J. Role of charged and hydrophobic residues in the oligomerization of the PYRIN domain of ASC. Biochemistry. 2005. 44:575–83.

37). Stehlik C., Krajewska M., Welsh K., Krajewski S., Godzik A., Reed JC. The PAAD/PYRIN-only protein POP1/ASC2 is a modulator of ASC-mediated nuclear-factor-kappa B and pro-caspase-1 regulation. Biochem J. 2003. 373:101–13.
38). Dorfleutner A., Bryan NB., Talbott SJ., Funya KN., Rellick SL., Reed JC, et al. Cellular pyrin domain-only protein 2 is a candidate regulator of inflammasome activation. Infect Immun. 2007. 75:1484–92.

39). Johnston JB., Barrett JW., Nazarian SH., Goodwin M., Ricciuto D., Wang G, et al. A poxvirus-encoded pyrin domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity. 2005. 23:587–98.

40). Fink SL., Bergsbaken T., Cookson BT. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc Natl Acad Sci U S A. 2008. 105:4312–7.

41). Masumoto J., Dowds TA., Schaner P., Chen FF., Ogura Y., Li M, et al. ASC is an activating adaptor for NF-kappa B and caspase-8-dependent apoptosis. Biochem Biophys Res Commun. 2003. 303:69–73.
42). Ohtsuka T., Ryu H., Minamishima YA., Macip S., Sagara J., Nakayama KI, et al. ASC is a Bax adaptor and regulates the p53-Bax mitochondrial apoptosis pathway. Nat Cell Biol. 2004. 6:121–8.

43). Yokoyama T., Sagara J., Guan X., Masumoto J., Takeoka M., Komiyama Y, et al. Methylation of ASC/TMS1, a proapoptotic gene responsible for activating procaspase-1, in human colorectal cancer. Cancer Lett. 2003. 202:101–8.

44). Lucas ME., Crider KS., Powell DR., Kapoor-Vazirani P., Vertino PM. Methylation-sensitive regulation of TMS1/ASC by the ETS factor, GA-binding protein-alpha. J Biol Chem. 2009. 284:14698–709.
45). Ting JP., Lovering RC., Alnemri ES., Bertin J., Boss JM., Davis BK, et al. The NLR gene family: a standard nomenclature. Immunity. 2008. 28:285–7.

46). Franchi L., Amer A., Body-Malapel M., Kanneganti TD., Ozören N., Jagirdar R, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006. 7:576–82.
47). Miao EA., Alpuche-Aranda CM., Dors M., Clark AE., Bader MW., Miller SI, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006. 7:569–75.
48). Suzuki T., Franchi L., Toma C., Ashida H., Ogawa M., Yoshikawa Y, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007. 3:e111.

49). Elinav E., Strowig T., Henao-Mejia J., Flavell RA. Regulation of the Antimicrobial Response by NLR Proteins. Immunity. 2011. 34:665–79.

50). Cui J., Zhu L., Xia X., Wang HY., Legras X., Hong J, et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010. 141:483–96.
51). Kumar H., Pandey S., Zou J., Kumagai Y., Takahashi K., Akira S, et al. NLRC5 deficiency does not influence cytokine induction by virus and bacteria infections. J Immunol. 2011. 186:994–1000.

52). Davis BK., Roberts RA., Huang MT., Willingham SB., Conti BJ., Brickey WJ, et al. Cutting edge: NLRC5-dependent activation of the inflammasome. J Immunol. 2011. 186:1333–7.

53). Faustin B., Lartigue L., Bruey JM., Luciano F., Sergienko E., Bailly-Maitre B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007. 25:713–24.

54). Boyden ED., Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006. 38:240–4.

55). Jin Y., Mailloux CM., Gowan K., Riccardi SL., LaBerge G., Bennett DC, et al. NALP1 in vitiligo-associated multiple autoimmune disease. N Engl J Med. 2007. 356:1216–25.
56). Halle A., Hornung V., Petzold GC., Stewart CR., Monks BG., Reinheckel T, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008. 9:857–65.
57). Dostert C., Pétrilli V., Van Bruggen R., Steele C., Mossman BT., Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008. 320:674–7.

58). Hornung V., Bauernfeind F., Halle A., Samstad EO., Kono H., Rock KL, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008. 9:847–56.

59). Chen GY., Nuñez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol. 2010. 10:826–37.

60). Duewell P., Kono H., Rayner KJ., Sirois CM., Vladimer G., Bauernfeind FG, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010. 464:1357–61.

61). Fernandes-Alnemri T., Yu JW., Juliana C., Solorzano L., Kang S., Wu J, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010. 11:385–93.
62). Zhou R., Yazdi AS., Menu P., Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011. 469:211–5.

63). Zhou R., Tardivel A., Thorens B., Choi I., Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010. 11:136–40.

64). Brydges SD., Mueller JL., McGeough MD., Pena CA., Misaghi A., Gandhi C, et al. Inflammasome-mediated disease animal models reveal roles for innate but not adaptive immunity. Immunity. 2009. 30:875–87.

65). Meng G., Zhang F., Fuss I., Kitani A., Strober W. A mutation in the Nlrp3 gene causing inflammasome hyperactivation potentiates Th17 cell-dominant immune responses. Immunity. 2009. 30:860–74.

66). Villani AC., Lemire M., Fortin G., Louis E., Silverberg MS., Collette C, et al. Common variants in the NLRP3 region contribute to Crohn's disease susceptibility. Nat Genet. 2009. 41:71–6.

67). Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. The International FMF Consortium. Cell. 1997. 90:797–807.
68). A candidate gene for familial Mediterranean fever. Nat Genet. 1997. 17:25–31.
69). Milhavet F., Cuisset L., Hoffman HM., Slim R., El-Shanti H., Aksentijevich I, et al. The infevers autoinflammatory mutation online registry: update with new genes and functions. Hum Mutat. 2008. 29:803–8.

70). Ozato K., Shin DM., Chang TH., Morse HC 3rd. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008. 8:849–60.

71). Uchil PD., Quinlan BD., Chan WT., Luna JM., Mothes W. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 2008. 4:e16.

72). Stremlau M., Owens CM., Perron MJ., Kiessling M., Autissier P., Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004. 427:848–53.
73). Gack MU., Shin YC., Joo CH., Urano T., Liang C., Sun L, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007. 446:916–20.

74). Tsuchida T., Zou J., Saitoh T., Kumar H., Abe T., Matsuura Y, et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010. 33:765–76.

75). Shi M., Deng W., Bi E., Mao K., Ji Y., Lin G, et al. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat Immunol. 2008. 9:369–77.
76). Zha J., Han KJ., Xu LG., He W., Zhou Q., Chen D, et al. The Ret finger protein inhibits signaling mediated by the noncanonical and canonical IkappaB kinase family members. J Immunol. 2006. 176:1072–80.
77). Richards N., Schaner P., Diaz A., Stuckey J., Shelden E., Wadhwa A, et al. Interaction between pyrin and the apoptotic speck protein (ASC) modulates ASC-induced apoptosis. J Biol Chem. 2001. 276:39320–9.

78). Dowds TA., Masumoto J., Chen FF., Ogura Y., Inohara N., Núñez G. Regulation of cryopyrin/Pypaf1 signaling by pyrin, the familial Mediterranean fever gene product. Biochem Biophys Res Commun. 2003. 302:575–80.

79). Chae JJ., Komarow HD., Cheng J., Wood G., Raben N., Liu PP, et al. Targeted disruption of pyrin, the FMF protein, causes heightened sensitivity to endotoxin and a defect in macrophage apoptosis. Mol Cell. 2003. 11:591–604.

80). Chae JJ., Wood G., Masters SL., Richard K., Park G., Smith BJ, et al. The B30.2 domain of pyrin, the familial Mediterranean fever protein, interacts directly with caspase-1 to modulate IL-1beta production. Proc Natl Acad Sci U S A. 2006. 103:9982–7.

81). Papin S., Cuenin S., Agostini L., Martinon F., Werner S., Beer HD, et al. The SPRY domain of Pyrin, mutated in familial Mediterranean fever patients, interacts with inflammasome components and inhibits proIL-1beta processing. Cell Death Differ. 2007. 14:1457–66.
82). Yu JW., Wu J., Zhang Z., Datta P., Ibrahimi I., Taniguchi S, et al. Cryopyrin and pyrin activate caspase-1, but not NF-kappaB, via ASC oligomerization. Cell Death Differ. 2006. 13:236–49.
83). Stehlik C., Lee SH., Dorfleutner A., Stassinopoulos A., Sagara J., Reed JC. Apoptosis-associated speck-like protein containing a caspase recruitment domain is a regulator of procaspase-1 activation. J Immunol. 2003. 171:6154–63.

84). Seshadri S., Duncan MD., Hart JM., Gavrilin MA., Wewers MD. Pyrin levels in human monocytes and monocyte-derived macrophages regulate IL-1beta processing and release. J Immunol. 2007. 179:1274–81.
85). Chae JJ., Cho YH., Lee GS., Cheng J., Liu PP., Feigenbaum L, et al. Gain-of-Function Pyrin Mutations Induce NLRP3 Protein-Independent Interleukin-1beta Activation and Severe Autoinflammation in Mice. Immunity. 2011. 34:755–68.
87). Tokunaga T., Yamamoto H., Shimada S., Abe H., Fukuda T., Fujisawa Y, et al. Antitumor activity of deoxyribonucleic acid fraction from Mycobacterium bovis BCG. I. Isolation, physicochemical characterization, and antitumor activity. J Natl Cancer Inst. 1984. 72:955–62.
88). Krieg AM., Yi AK., Matson S., Waldschmidt TJ., Bishop GA., Teasdale R, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995. 374:546–9.

89). Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000. 408:740–5.

90). Ishii KJ., Suzuki K., Coban C., Takeshita F., Itoh Y., Matoba H, et al. Genomic DNA released by dying cells induces the maturation of APCs. J Immunol. 2001. 167:2602–7.

91). Yasuda K., Yu P., Kirschning CJ., Schlatter B., Schmitz F., Heit A, et al. Endosomal translocation of vertebrate DNA activates dendritic cells via TLR9-dependent and -independent pathways. J Immunol. 2005. 174:6129–36.

92). Takaoka A., Wang Z., Choi MK., Yanai H., Negishi H., Ban T, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007. 448:501–5.

93). Ishii KJ., Kawagoe T., Koyama S., Matsui K., Kumar H., Kawai T, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008. 451:725–9.

94). Ishikawa H., Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008. 455:674–8.

95). Zhong B., Yang Y., Li S., Wang YY., Li Y., Diao F, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008. 29:538–50.

96). Ishikawa H., Ma Z., Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009. 461:788–92.

97). Yanai H., Ban T., Wang Z., Choi MK., Kawamura T., Negishi H, et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009. 462:99–103.

98). Unterholzner L., Keating SE., Baran M., Horan KA., Jensen SB., Sharma S, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010. 11:997–1004.

99). Muruve DA., Pétrilli V., Zaiss AK., White LR., Clark SA., Ross PJ, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008. 452:103–7.

100). Albrecht M., Choubey D., Lengauer T. The HIN domain of IFI-200 proteins consists of two OB folds. Biochem Biophys Res Commun. 2005. 327:679–87.

101). Ludlow LE., Johnstone RW., Clarke CJ. The HIN-200 family: more than interferon-inducible genes? Exp Cell Res. 2005. 308:1–17.

102). Rathinam VA., Jiang Z., Waggoner SN., Sharma S., Cole LE., Waggoner L, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010. 11:395–402.

103). Jones JW., Kayagaki N., Broz P., Henry T., Newton K., O'Rourke K, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci U S A. 2010. 107:9771–6.
104). Henry T., Brotcke A., Weiss DS., Thompson LJ., Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007. 204:987–94.

105). Woerner SM., Kloor M., Schwitalle Y., Youmans H., Doeberitz MK., Gebert J, et al. The putative tumor suppressor AIM2 is frequently affected by different genetic alterations in microsatellite unstable colon cancers. Genes Chromosomes Cancer. 2007. 46:1080–9.
Go to : 
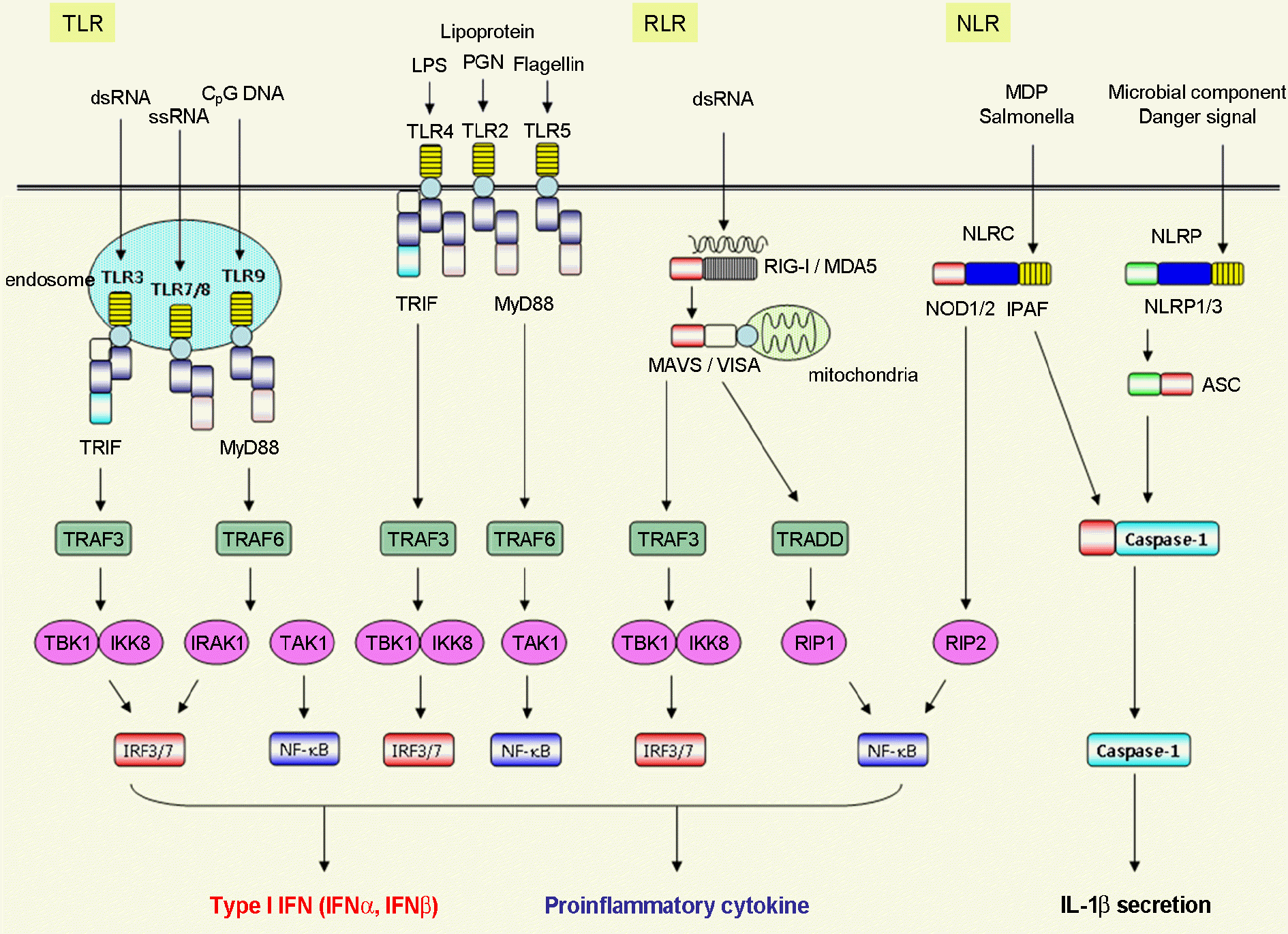 | Figure 1.
Pattern-recognition receptor (PRR) signaling pathways. Three classes of PRR families, such as TLRs, RLRs and NLRs, sense microbial products or endogenous danger signals to trigger downstream signaling pathways, resulting in the production of type I IFN, proinflammatory cytokines and the activation of caspase-1. |
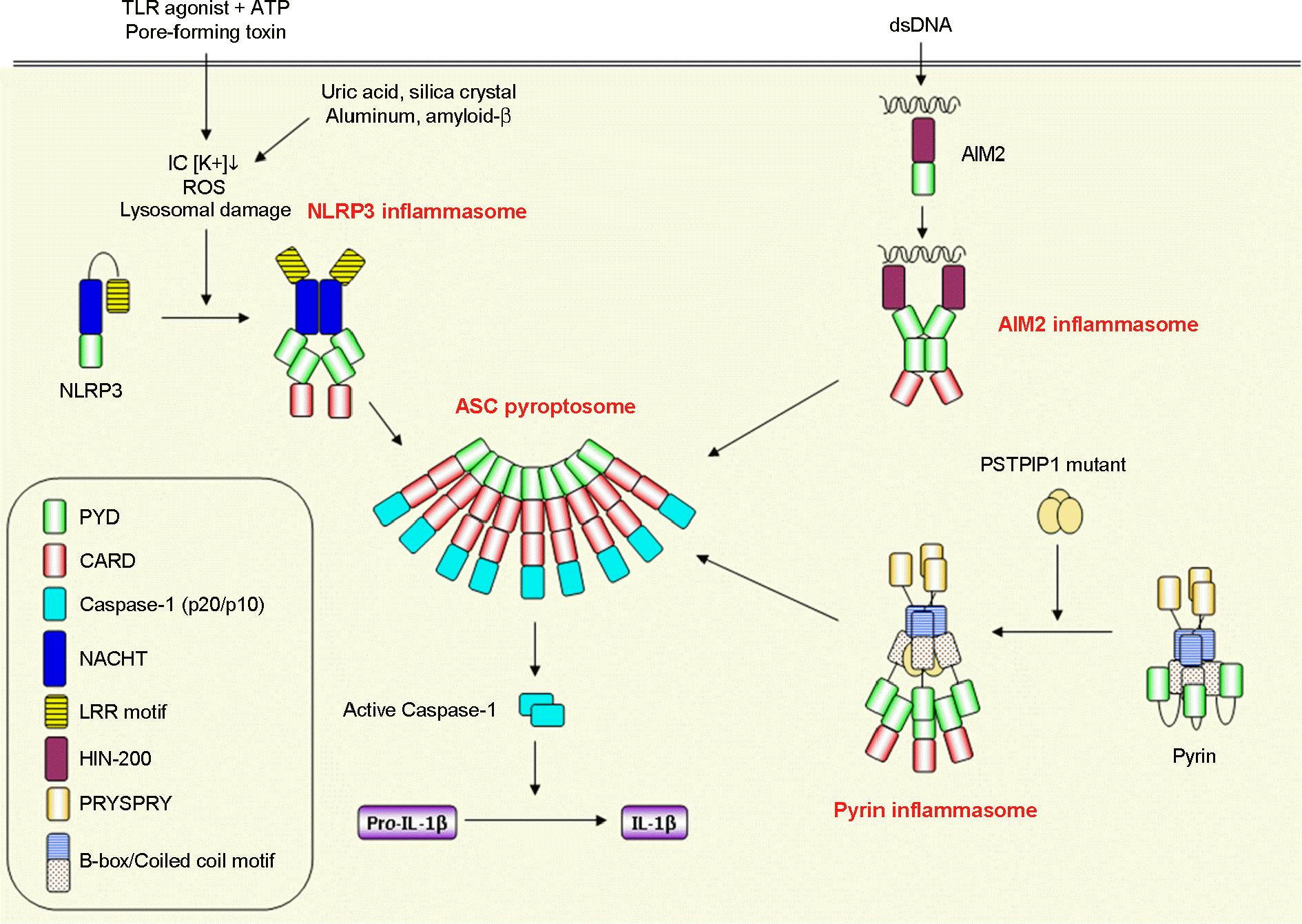 | Figure 2.
PYD-containing inflammasome pathways. Inactive PYD-containing inflammasome components are activated by sensing diverse microbial infection or danger signals (NLRP3), cytoplasmic dsDNA (AIM2) and autoinflammatory disease-associated PSTPIP1 mutant (pyrin) to trigger the formation of ASC pyroptosome, resulting in the activation of caspase-1. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download