Abstract
Turnip yellow mosaic virus (TYMV) is a spherical plant virus that has a single 6.3 kb positive strand RNA genome. Information for TYMV replication is limited, except that the 3′-terminal sequence and 5′-untranslated region are required for genome replication. When a foreign sequence was inserted at the position upstream of the coat protein (CP) open reading frame (ORF), replication of the recombinant TYMV was comparable to wild type, as long as an RNAi suppressor was provided. In contrast, when the foreign sequence was inserted between the CP ORF and the 3′-terminal tRNA-like structure, replication of the recombinant virus was not detected. This result suggests that the CP ORF contains an essential replication element which should be appropriately spaced with respect to the 3′-end. Analysis of TYMV constructs containing a part or a full additional CP ORF indicates that the 3′ quarter of the CP ORF is required for TYMV replication.
Go to : 
REFERENCES
1). Dreher TW. Turnip yellow mosaic virus: transfer RNA mimicry, chloroplasts and a C-rich genome. Mol Plant Pathol. 2004. 5:367–75.

2). Bozarth CS., Weiland JJ., Dreher TW. Expression of ORF-69 of turnip yellow mosaic virus is necessary for viral spread in plants. Virology. 1992. 187:124–30.

3). Tsai CH., Dreher TW. Increased viral yield and symptom severity result from a single amino acid substitution in the turnip yellow mosaic virus movement protein. Mol Plant Microbe Interact. 1993. 6:268–73.

4). Chen J., Li WX., Xie D., Peng JR., Ding SW. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell. 2004. 16:1302–13.
5). Dreher TW., Goodwin JB. Transfer RNA mimicry among tymoviral genomic RNAs ranges from highly efficient to vestigial. Nucleic Acids Res. 1998. 26:4356–64.

6). Dreher TW., Tsai CH., Florentz C., Giegé R. Specific valylation of turnip yellow mosaic virus RNA by wheat germ valyltRNA synthetase determined by three anticodon loop nucleotides. Biochemistry. 1992. 31:9183–9.

7). Matsuda D., Dreher TW. The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology. 2004. 321:36–46.

8). Miller WA., Bujarski JJ., Dreher TW., Hall TC. Minus-strand initiation by brome mosaic virus replicase within the 3′ tRNA-like structure of native and modified RNA templates. J Mol Biol. 1986. 187:537–46.

9). Deiman BA., Koenen AK., Verlaan PW., Pleij CW. Minimal template requirements for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J Virol. 1998. 72:3965–72.
10). Shin HI., Cho NJ., Cho TJ. Role of 5′-UTR hairpins of the Turnip yellow mosaic virus RNA in replication and systemic movement. BMB Rep. 2008. 41:778–83.

11). Shin HI., Tzanetakis IE., Dreher TW., Cho TJ. The 5′-UTR of Turnip yellow mosaic virus does not include a critical encapsidation signal. Virology. 2009. 387:427–35.

12). Shin HI., Kim IC., Cho TJ. Replication and encapsidation of recombinant Turnip yellow mosaic virus RNA. BMB Rep. 2008. 41:739–44.

13). Cho TJ., Dreher TW. Encapsidation of genomic but not sub-genomic turnip yellow mosaic virus RNA by coat protein provided in trans. Virology. 2006. 356:126–35.

14). Voinnet O., Pinto YM., Baulcombe DC. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci. 1999. 96:14147–52.

15). Ray D., White KA. Enhancer-like properties of an RNA element that modulates tombusvirus RNA accumulation. Virology. 1999. 256:162–71.

16). White KA., Nagy PD. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog Nucleic Acid Res Mol Biol. 2004. 78:187–226.

17). Pogany J., Fabian MR., White KA., Nagy PD. A replication silencer element in a plus-strand RNA virus. EMBO J. 2003. 22:5602–11.

18). Ray D., Na H., White KA. Structure properties of a multifunctional T-shaped RNA domain that mediate efficient Tomato bushy stunt virus RNA replication. J Virol. 2004. 78:10490–500.
19). Panaviene Z., Panavas T., Nagy PD. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J Virol. 2005. 79:10608–18.

Go to : 
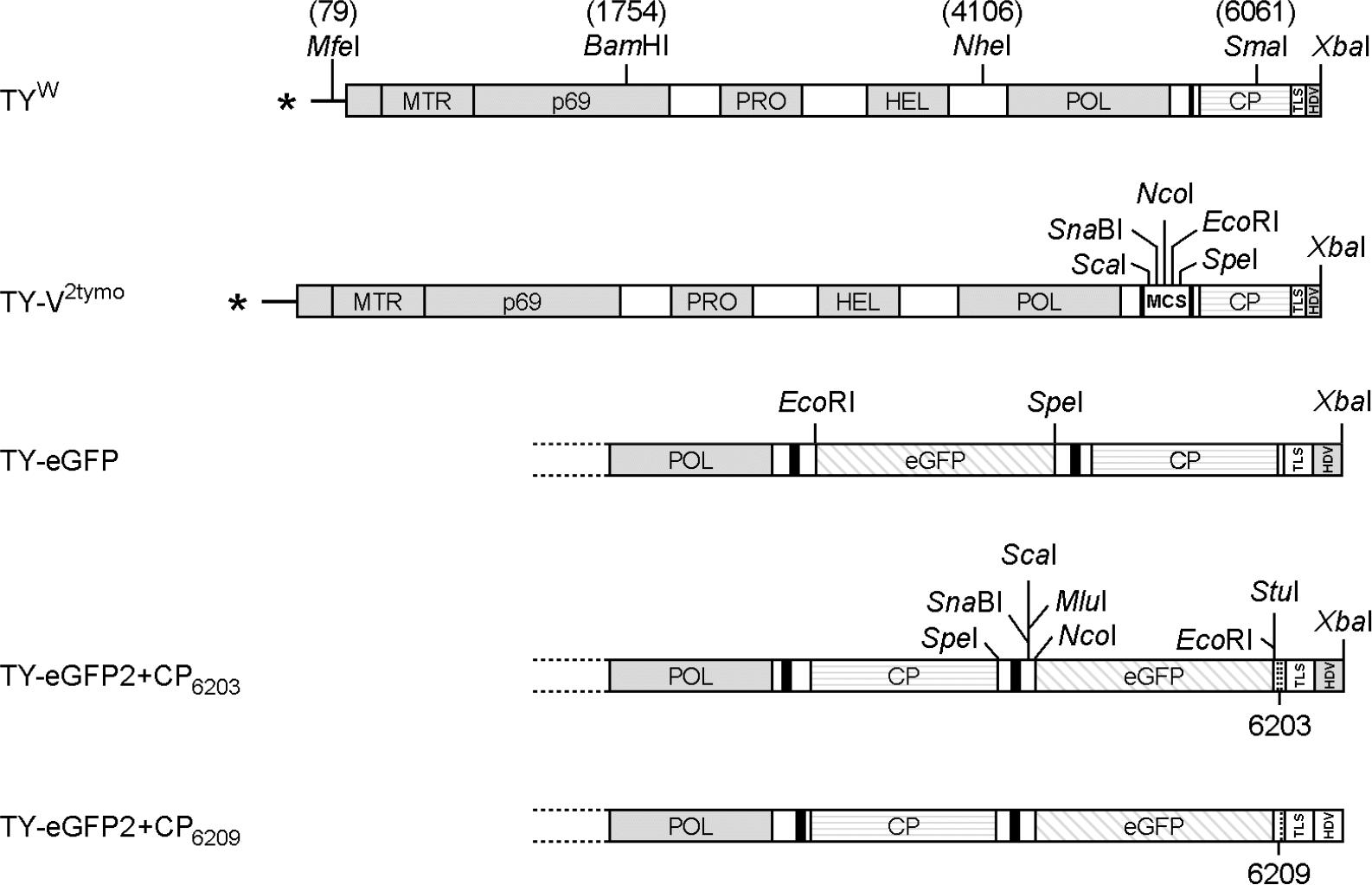 | Figure 1.
Recombinant TYMV constructs. TYW represents a wild type genome. TY-V2tymo is an expression vector where an extra tymobox and a multiple cloning site were inserted between the pol domain and the coat protein (CP) ORF. The gene encoding an enhanced green fluorescent protein (eGFP) was cloned into the TY-V2tymo, yielding TY-eGFP. In the TY-eGFP construct, the eGFP gene is expressed as a bigger sgRNA. TY-eGFP2 constructs were designed to have the eGFP gene between the CP ORF and the 3′-terminal TLS. The eGFP gene here is expressed as a smaller sgRNA. In the TY-eGFP2+CP6203 and TY-eGFP2+CP6209 constructs, the TYMV sequence downstream of the eGFP gene begins with the nucleotide at nt-6203 and nt-6209 of the wild type, respectively. All these constructs have a 3′-terminal ribozyme sequence derived from hepatitis delta virus (HDV). |
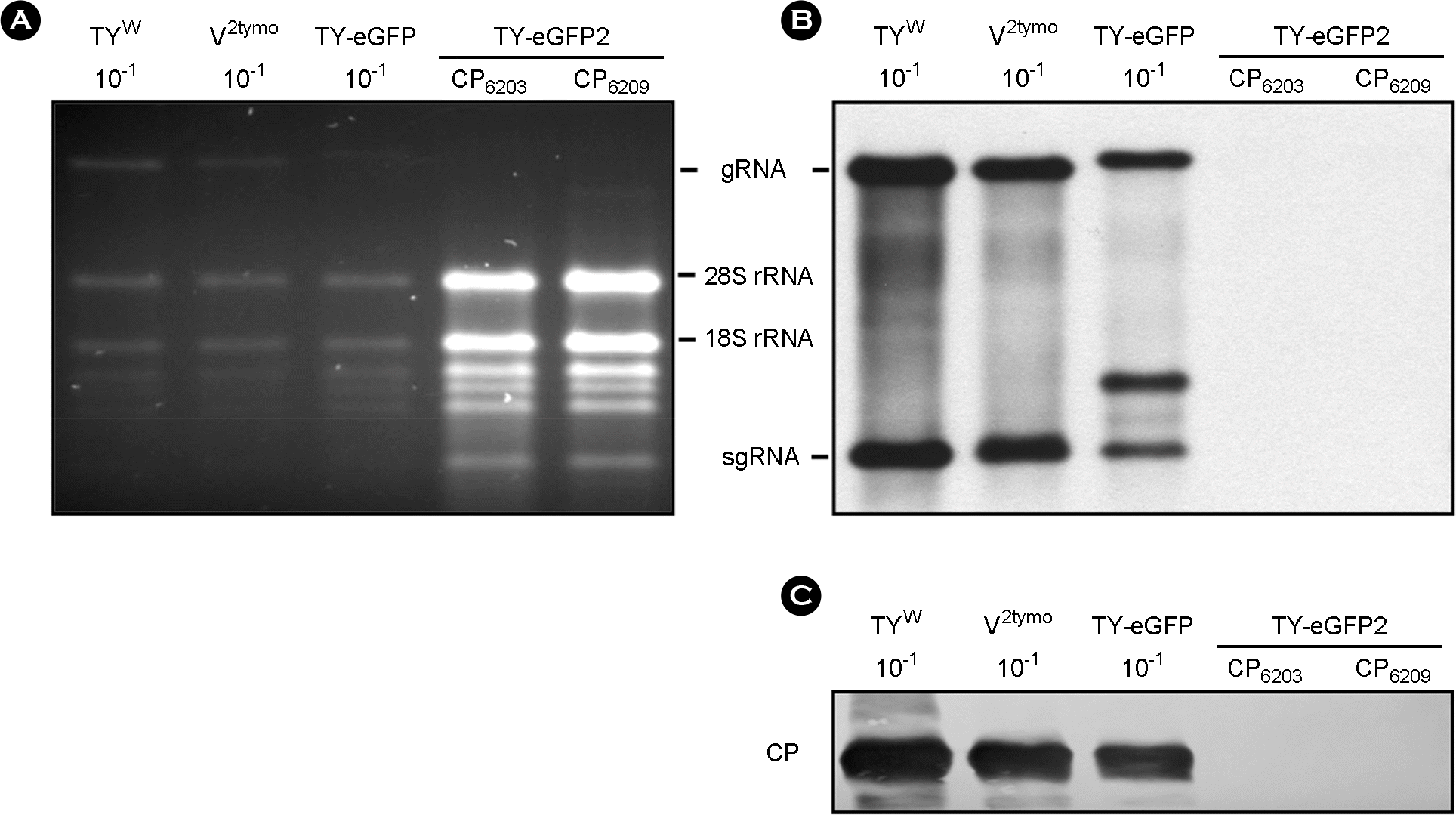 | Figure 2.
Replication of TY-eGFP and TY-eGFP2 in N. benthamiana. Seven days after agroinfiltration of N. benthamiana leaf with various TYMV constructs, total RNA was extracted from the leaf. 1 μg or 0.1 μg (10–1) of total RNA was size-fractionated in the 1% agarose gel (A) and examined by Northern blot analysis (B), using the DIG-labeled probe representing the CP ORF. (C) Western analysis of coat protein expression. 1:10 diluted (10–1) or undiluted leaf extract was loaded and electrophoresed in 12.5% SDS-polyacrylamide gel. The proteins were transferred to a nitrocellulose membrane. Coat protein was detected using anti-TYMV coat protein rabbit antibody and anti-rabbit HRP conjugate. The membrane was developed by a Luminata™ Forte (Millipore) using luminol as the substrate. The results of the Northern and Western analyses show that the replication of the TY-eGFP2 is impaired, in contrast to the TY-eGFP. |
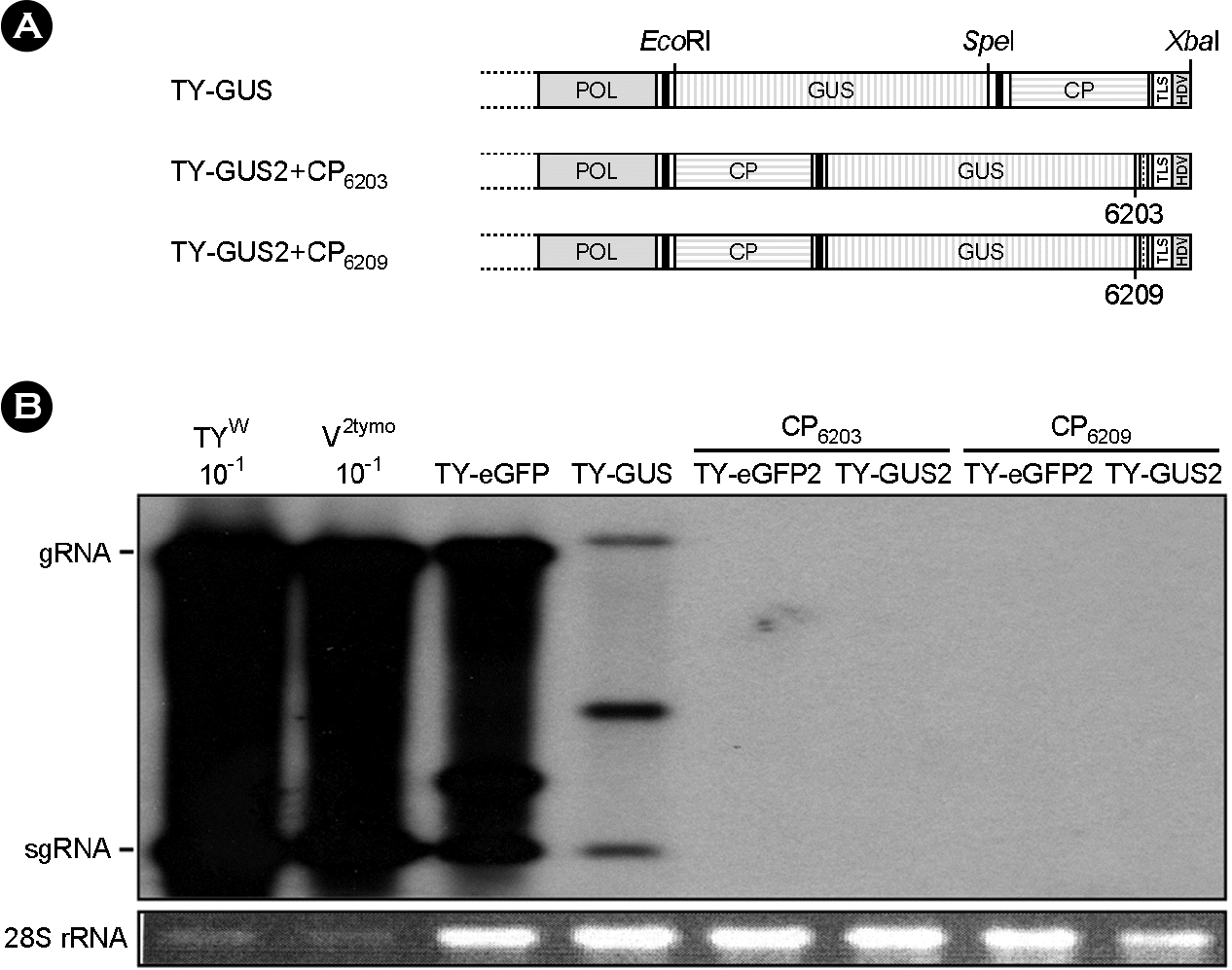 | Figure 3.
Replication of recombinant TYMV constructs in Chinese cabbage. (A) TY-GUS and TY-GUS2 constructs. (B) Analysis of recombinant TYMV replication. TY-eGFP, TY-eGFP2, TY-GUS, and TY-GUS2 constructs were inoculated into Chinese cabbage. Northern blot analysis was done as described in Figure 2. The results show that the recombinant TYMV constructs do not replicate well either in the natural host when they have an extra sequence between the CP ORF and the 3′-TLS. |
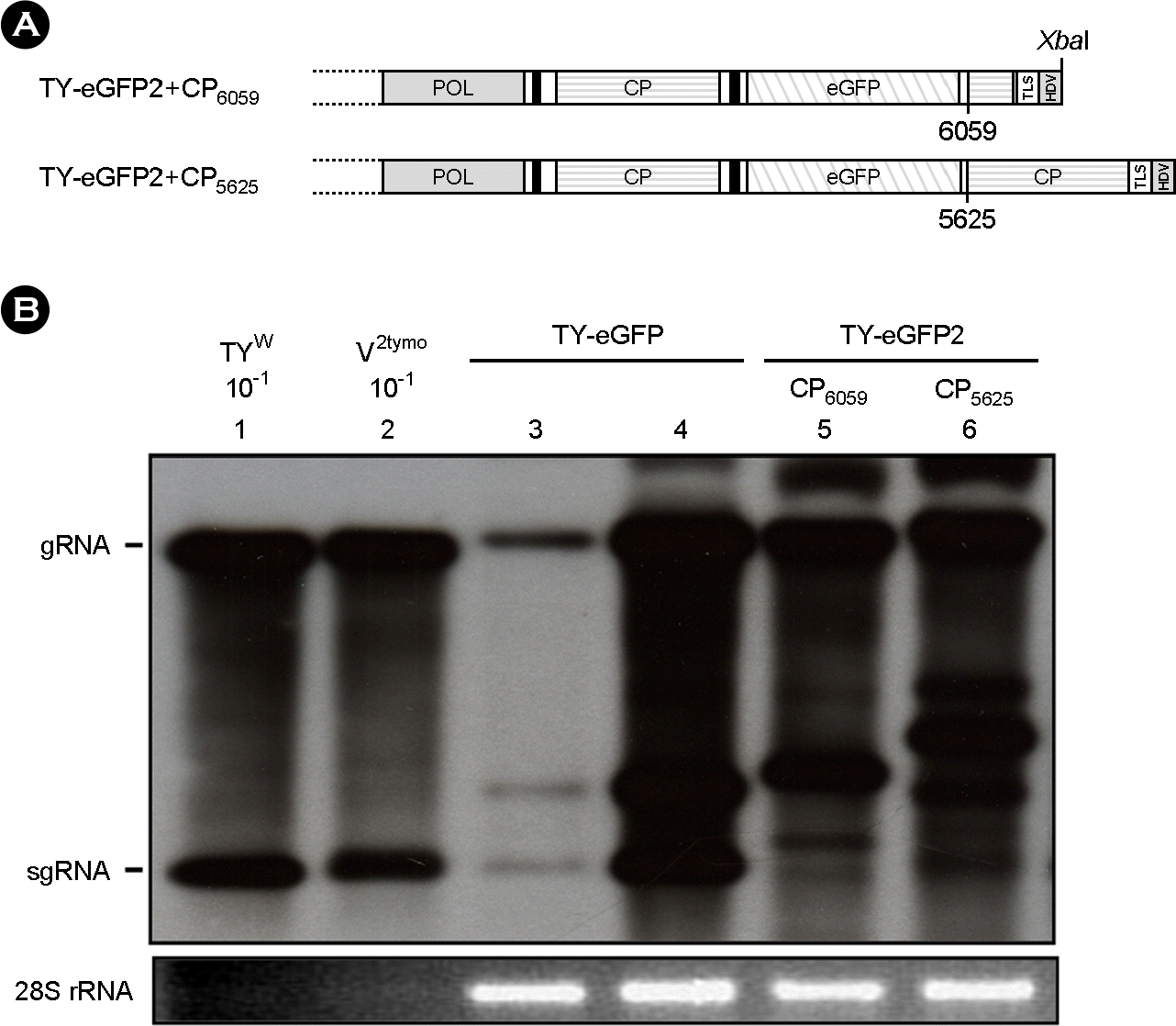 | Figure 4.
Replication of TY-eGFP2 constructs with an additional CP ORF sequence. (A) TY-eGFP2+CP6059 and TY-eGFP2+CP5625 constructs. TY-eGFP2+ CP6059 contains an additional 3′-quarter of the CP ORF at the downstream of the eGFP. TY-eGFP2+CP5625 has a whole additional CP ORF sequence after the eGFP. (B) Northern analysis of the recombinant TYMV replication. All agroinfiltrations included expression of the p19 RNAi suppressor except for the inoculation represented by lane 3. Northern analysis was done as described in Figure 2. The results shown here indicate that a sequence element essential for TYMV replication resides in the 3′-quarter of the CP ORF. |
Table 1.
Oligonucleotides used in this study
| Primer name | Sequence∗ |
|---|---|
| TY(+)6209 | 5′-CGAGAATTC AGGCCTAAGTTCTCGATC-3′ |
| TY(+)6186 | 5′-CGAGAATTC AGGCCTTCCGCTCATCACGGACACTTCCACCT-3′ |
| TY(+)6139 | 5′-CGAGAATTC AGGCCTGCATCGACCTGCATAATAACTGTAT-3′ |
| TY(+)6059 | 5′-CGAGAATTC AGGCCTCCCGGGTCAAAGATTCGATTCAGT-3′ |
| TY(+)5625 | 5′-CGAGAATTC AGGCCTAATAGCAATCAGCCCCAACAT-3′ |
| HDV-R29 | 5′-GGAGAATTC TCTAGATGGCTCTCCCTTAG-3′ |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download