Abstract
Cryptocococcus neoformans is an encapsulated yeast that can cause life-threatening infections in immunocompromised patients. In this study, the genetic variability and epidemiological relationships of clinical and environmental isolates of C. neoformans from Busan, Korea, 2000~2005 were investigated. A total of 12 strains of C. neoformans, 7 clinical and 5 environmental isolates were analyzed by random amplified polymorphic DNA (RAPD) using three different primers and PCR-fingerprinting with a minisatellite-specific core sequence of phage M13. All strains belonged to C. neoformans serotype A and mating type MATa. Two different RAPD profiles (I and II) and a single pattern by M13 PCR-fingerprinting were identified. The major RAPD profile was pattern I (8 of 12 strains) and pattern II was identified from 2 clinical and 2 environmental strains, which clearly distinguished among isolates. Clinical strains with pattern II were isolated from the patients with HIV positive. Taken together, molecular patterns provide a good characterization of strains of C. neoformans as a heterogeneous group and epidemiological relationships in clinical and environmental strains.
REFERENCES
1). Matsumoto MT., Fusco-Almeida AM., Baeza LC., Melhem Mde S., Medes-Giannini MJ. Genotyping, serotyping and determination of mating-type of Cryptococcus neoformans clinical isolates from São Paulo State, Brazil. Rev Inst Med Trop Sao Paulo. 2007. 49:41–7.
2). Chen J., Varma A., Diaz MR., Litvintseva AP., Wollenberg KK., Kwon-Chung KJ. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg Infect Dis. 2008. 14:755–62.
3). Feng X., Yao Z., Ren D., Liao W., Wu J. Genotype and mating type analysis of Cryptococcus neoformans and Cryptococcus gattii isolates from China that mainly originated from non-HIV-infected patients. FEMS Yeast Res. 2008. 8:930–8.
4). Benett JE., Kwon-Chung KJ., Howard DH. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am J Epidermiol. 1977. 105:582–6.
5). Walter JE., Coffee EG. Distribution and epidemiologic significance of the serotypes of Cryptococcus neoformans. Am J Epidemiol. 1968. 87:167–72.
6). Franzot SP., Hamdan JS., Currie BP., Casadevall A. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: Evidence for both local genetic differences and a global clonal population structure. J Clin Microbiol. 1997. 35:2243–51.
7). Pfeiffer TJ., Ellis DH. Environmental isolation of Cryptococcus neoformans var. gattii from Eucalyptus tereticornis. J Med Vet Mycol. 1992. 30:407–8.
8). Ellis DH., Pfeiffer TJ. Natural habitat of Cryptococcus neoformans var. gattii. J Clin Microbiol. 1990. 28:1642–4.
9). Kuroki M., Phichaichumpon C., Yasuoka A., Chiranairadul P., Chosa T., Sirinirund P, et al. Environmental isolation of Cryptococcus neoformans from endemic region of HIV-associated cryptococcosis in Thailand. Yeast. 2004. 21:809–12.
10). Ito-Kuwa S., Nagamura K., Aoki S., Ninimiya K., Kato J., Vidotto V. Serotyping of Cryptococccus neoformans isolated from AIDS patients. Shigaku (Odontology). 1994. 82:360–4.
11). Trilles L., Lazéra Mdos S., Wanke B., Oliveira RV., Barbosa GG., Nishikawa MM, et al. Regional pattern of the molecular types of Cryptococcus neoformans and Cryptococcus gattii in Brazil. Mem Inst Oswaldo Cruz. 2008. 103:455–62.
12). Chaturvedi S., Rodeghier B., Fan J., McClelland CM., Wickes BL., Chaturvedi V. Direct PCR of Cryptococcus neoformans MATalpha and MATa pheromones to determine mating type, ploidy, and variety: a tool for epidemiological and molecular pathogenesis studies. J Clin Microbiol. 2000. 38:2007–9.
13). Jain N., Wickes BL., Keller SM., Fu J., Casadevall A., Jain P, et al. Molecular epidemiology of clinical Cryptococcus neoformans strains from India. J Clin Microbiol. 2005. 43:5733–42.
14). Meyer W., Marszewska K., Amirmostofian M., Igreja RP., Hardtke C., Methling K, et al. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA - a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis. 1999. 20:1790–9.
15). Boekhout T., Theelen B., Diaz M., Fell JW., Hop WC., Abeln EC, et al. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology. 2001. 147:891–907.
16). Diaz MR., Boekhout T., Kiesling T., Fell JW. Comparative analysis of the intergenic spacer regions and population structure of the species complex of the pathogenic yeast Cryptococcus neoformans. FEMS Yeast Res. 2005. 5:1129–40.
17). Brandt ME., Hutwagner LC., Kuykendall RJ., Pinner RW. Comparison of multilocus enzyme electrophoresis and random amplified polymorphic DNA analysis for molecular subtyping of Cryptococcus neoformans. The Cryplococcal Disease Active Surveillance Group. J Clin Microbiol. 1995. 33:1890–5.
18). Litvintseva AP., Thakur R., Vilgalys R., Mitchell TG. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics. 2006. 172:2223–38.
19). Chung SM., Lee EY., Lee CK., Eom DW., Yoo B., Moon HB. A case of cryptococcal tenosynovitis in a patient with Wegener's granulomatosis. J Kor Rheumatism Ass. 2004. 11:66–71.
20). Oh KS., Hwang SM. Isolation and characterization of Cryptococcus neoformans from environmental sources in Busan. Mycobiology. 2005. 33:188–93.
21). Kwon-Chung KJ., Polacheck I., Bennett JE. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotype B and C). J Clin Microbiol. 1982. 15:535–7.
22). Kwon-Chung KJ., Wickes BL., Booth JL., Vishniac HS., Bennett JE. Urease inhibition by EDTA in the two varieties of Cryptococcus neoformans. Infect Immun. 1987. 55:1751–4.
23). Yamamoto Y., Kohno S., Koga H., Kakeya H., Tomono K., Kaku M, et al. Random amplified polymorphic DNA analysis of clinically and environmentally isolated Cryptococcus neoformans in Nagasaki. J Clin Microbiol. 1995. 33:3328–32.
24). Ohkusu M., Tangonan N., Takeo K., Kishida E., Ohkubo M., Aoki S, et al. Serotype, mating type and ploidy of Cryptococcus neoformans strains isolated from patients in Brazil. Rev Inst Med Trop Sao Paulo. 2002. 44:299–302.
25). Kim SJ., Kim SO., Lee SH., Chong YS., Suk JS. A study on the mating types and serotypes of clinical isolates of Cryptococcus neoformans and production of serodiagnostic antigen and antiserum for cryptococcosis. Korean J Microbiol. 1986. 21:127–31.
26). Hwang SM., Oh KS., Lee KW. Serotype and enzymatic profile of Cryptococcus neoformans isolates from clinical and environmental sources in Korea. Korean J Microbiol. 2006. 42:257–64.
27). Ikeda R., Shinoda T., Fukazawa Y., Kaufman L. Antigenic characterization of Cryptococcus neoformans serotypes and its application to serotyping of clinical isolates. J Clin Microbiol. 1982. 16:22–9.
28). Kwon-Chung KJ., Bennett JF. Distribution of alpha and alpha mating types of Cryptococcus neoformans among natural and clinical isolates. Am J Epidemiol. 1978. 108:337–40.
29). Meyer W., Castañeda A., Jackson S., Huynh M., Castañeda E. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis. 2003. 9:189–95.
30). Boekhout T., van Belkum A. Variability of karyotypes and RAPD types in genetically related strains of Cryptococcus neoformans. Curr Genet. 1997. 32:203–8.
Figure 1.
PCR products of C. neoformans isolates for determination of mating type α and a. Lane 1~5: clinical and environmental isolates, 6: negative control, M: molecular marker
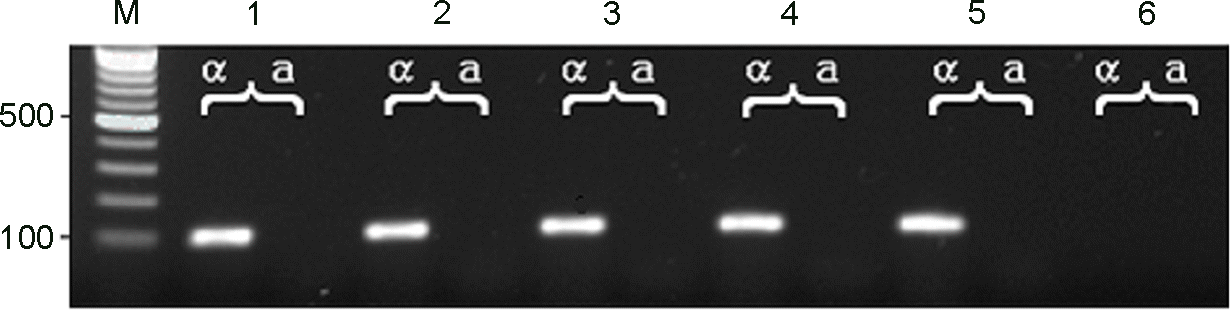
Figure 2.
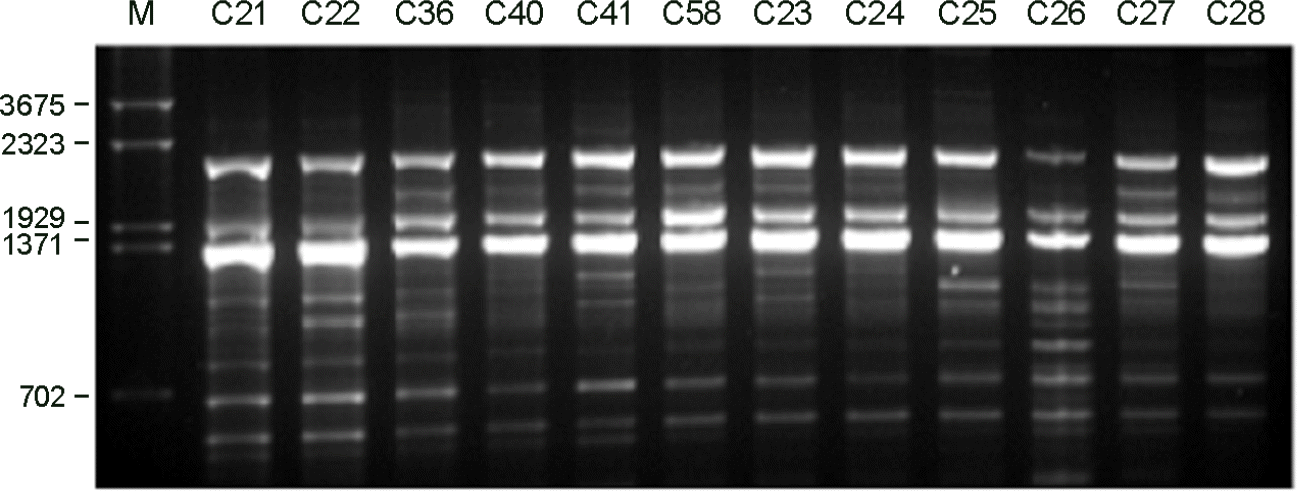
RAPD profiles of C. neoformans isolates amplified with the primers. (A): Primer OPH-02, (B): Primer OPH-12, (C): Primer OPH-20. M: molecular marker.
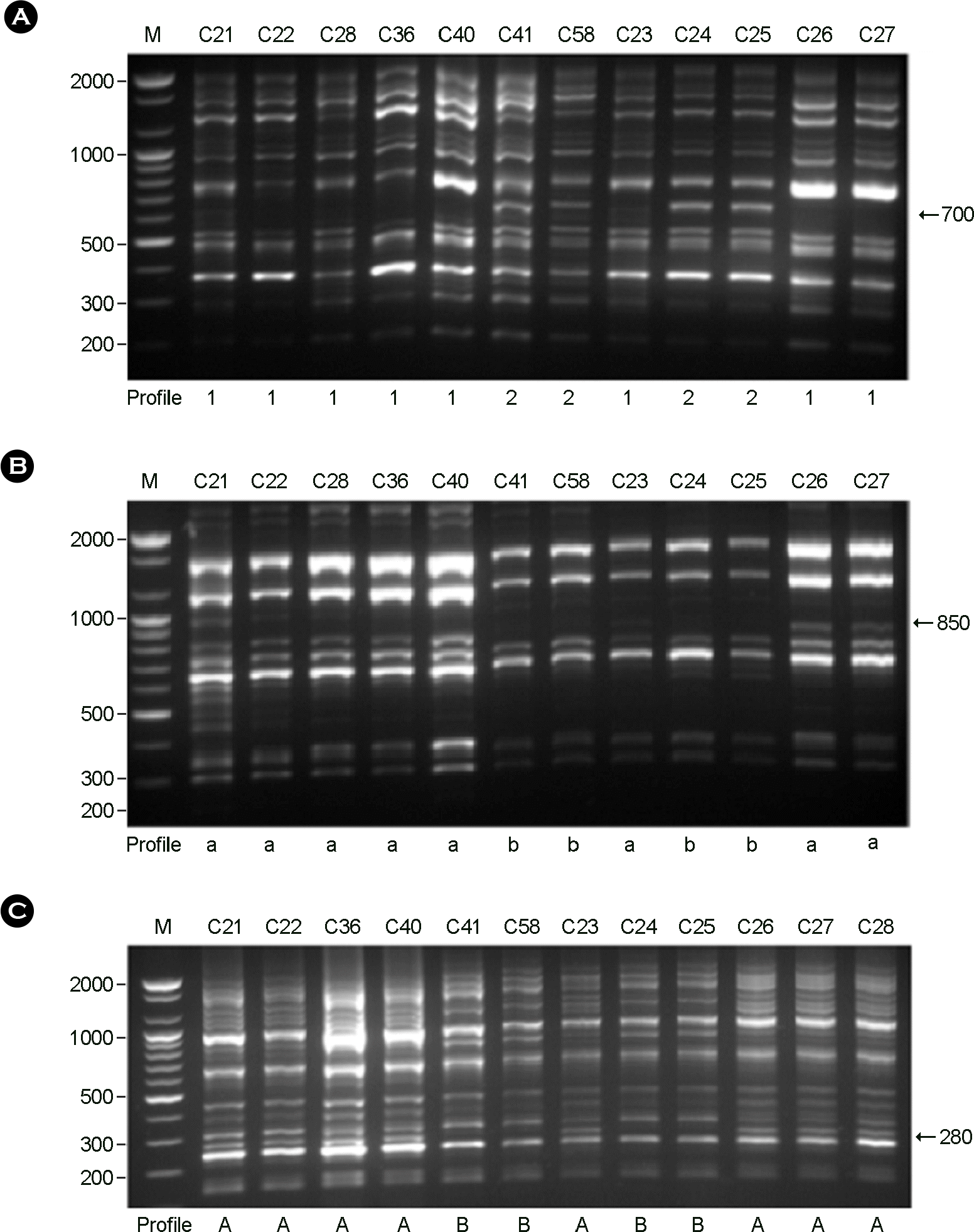
Table 1.
Sources, serotypes, and mating types of C. neoformans isolates in Busan, Korea
| No. | Strain | Type of isolates | Source | Year of isolation | Serotype | Mating type |
|---|---|---|---|---|---|---|
| 1 | C21 | aC | CSF | 2000 | A | a |
| 2 | C22 | C | CSF | 2001 | A | a |
| 3 | C28 | C | Blood | 2002 | A | a |
| 4 | C36 | C | Blood | 2003 | A | a |
| 5 | C40 | C | cCSF | 2003 | A | a |
| 6 | C41 | C | Blood | 2005 | A | a |
| 7 | C58 | C | CSF | 2004 | A | a |
| 8 | C23 | bE | Pigeon excreta | 2002 | A | a |
| 9 | C24 | E | Pigeon excreta | 2002 | A | a |
| 10 | C25 | E | Pigeon excreta | 2002 | A | a |
| 11 | C26 | E | Pigeon excreta | 2002 | A | a |
| 12 | C27 | E | Pigeon excreta | 2002 | A | a |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download