Abstract
Bovine coronavirus (BCoV) causes severe diarrhea in newborn calves, and is associated with winter dysentery in adult cattle and respiratory infections in calves and feedlot cattle. Although the Korean BCoV vaccine strain, BC94, was isolated in 1995, there has still been no report of a molecular characterization of the vaccine strain. To characterize the vaccine strain, relationships between BC94 and field strains were investigated, based on sequence analysis and cross-immunity. We determined the complete sequences of the HE, N, and S genes from BC94 and four NVRQS isolates (SUN5, A3, 0501, 0502). Due to its major role in antigenicity, the spike proteins of the BCoVs were analyzed. BC94 showed distinctive genetic divergence from field isolates collected from 2002 to 2005. BC94, SUN5, and A3 had no virulence-specific sequence and there was a single amino acid change, from asparagine to lysine at residue 175, in the polymorphic region. Strains 0501 and 0502 had virulence-specific sequences at all seven sites. Although the recently isolated Korean BCoVs and BC94 were genetically different, the cleavage site of spike genes at 763~768 (KRRSRR) and the antigenic domain II of the spike protein, amino acid position 528, were conserved in all NVRQS isolates. The antigenic relatedness of KCD9, representative of recent Korean BCoVs, was compared with the Korean vaccine strain BC94. KCD9 showed cross-reactivity against BC94 by virus neutralization (VN) test. These results suggest that BC94 is antigenically closely related to field isolates and is still effective as a vaccine strain.
Go to : 
REFERENCES
1). Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997. 142:629–33.
2). Cho KO., Halbur PG., Bruna JD., Sorden SD., Yoon KJ., Janke BH., Chang KO., Saif LJ. Detection and isolation of coronavirus from feces of three herds of feedlot cattle during outbreaks of winter dysentery-like disease. J Am Vet Med Assoc. 2000. 217:1191–4.

3). Clark MA. Bovine coronavirus. Br Vet J. 1993. 149:51–70.
4). Lathrop SL., Wittum TE., Brock KV., Loerch SC., Perino LJ., Bingham HR., McCollum FT., Saif LJ. Association between infection of the respiratory tract attributable to bovine coronavirus and health and growth performance of cattle in feedlots. Am J Vet Res. 2000. 61:1062–6.

5). Saif LJ., Brock KV., Redman DR., Kohler EM. Winter dysentery in dairy herds: electron microscopic and serological evidence for an association with coronavirus infection. Vet Rec. 1991. 128:447–9.

6). Storz J., Purdy CW., Lin X., Burrell M., Truax RE., Briggs RE., Frank GH., Loan RW. Isolation of respiratory bovine coronavirus, other cytocidal viruses, and Pasteurella spp from cattle involved in two natural outbreaks of shipping fever. J Am Vet Med Assoc. 2000. 216:1599–604.
8). Schultze B., Gross HJ., Brossmer R., Herrler G. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J Virol. 1991. 65:6232–7.

9). Schultze B., Wahn K., Klenk HD., Herrler G. Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-destroying and receptor-binding activity. Virology. 1991. 180:221–8.

10). Cavanagh D., Davis PJ. Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J Gen Virol. 1986. 67:1443–8.

11). Saeki K., Ohtsuka N., Taguchi F. Identification of spike protein residues of murine coronavirus responsible for receptor-binding activity by use of soluble receptor-resistant mutants. J Virol. 1997. 71:9024–31.

12). Yoo D., Parker MD., Babiuk LA. Analysis of the S spike (peplomer) glycoprotein of bovine coronavirus synthesized in insect cells. Virology. 1990. 179:121–8.

13). Storz J., Rott R., Kaluza G. Enhancement of plaque formation and cell fusion of an enteropathogenic coronavirus by trypsin treatment. Infect Immun. 1981. 31:1214–22.

14). Sturman LS., Ricard CS., Holmes KV. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: activation of cell-fusing activity of virions by trypsin and separation of two different 90K cleavage fragments. J Virol. 1985. 56:904–11.

15). Deregt D., Babiuk LA. Monoclonal antibodies to bovine coronavirus: characteristics and topographical mapping of neutralizing epitopes on the E2 and E3 glycoproteins. Virology. 1987. 161:410–20.

16). Grosse B., Siddell SG. Single amino acid changes in the S2 subunit of the MHV surface glycoprotein confer resistance to neutralization by S1 subunit-specific monoclonal antibody. Virology. 1994. 202:814–24.

17). Luytjes W., Geerts D., Posthumus W., Meloen R., Spaan W. Amino acid sequence of a conserved neutralizing epitope of murine coronaviruses. J Virol. 1989. 63:1408–12.

18). Chouljenko VN., Kousoulas KG., Lin X., Storz J. Nucleotide and predicted amino acid sequences of all genes encoded by the 3′ genomic portion (9.5 kb) of respiratory bovine coronaviruses and comparisons among respiratory and enteric coronaviruses. Virus Genes. 1998. 17:33–42.
19). Zuo X., Xiao Y., Plaxco KW. High specificity, electrochemical sandwich assays based on single aptamer sequences and suitable for the direct detection of small-molecule targets in blood and other complex matrices. J Am Chem Soc. 2009. 131:6944–5.

20). Lin XQ., KL Oe., Storz J., Purdy CW., Loan RW. Antibody responses to respiratory coronavirus infections of cattle during shipping fever pathogenesis. Arch Virol. 2000. 145:2335–49.

21). Yoo D., Deregt D. A single amino acid change within antigenic domain II of the spike protein of bovine coronavirus confers resistance to virus neutralization. Clin Diagn Lab Immunol. 2001. 8:297–302.

22). Abraham S., Kienzle TE., Lapps W., Brian DA. Deduced sequence of the bovine coronavirus spike protein and identification of the internal proteolytic cleavage site. Virology. 1990. 176:296–301.

23). Boireau P., Cruciere C., Laporte J. Nucleotide sequence of the glycoprotein S gene of bovine enteric coronavirus and comparison with the S proteins of two mouse hepatitis virus strains. J Gen Virol. 1990. 71:487–92.

24). Storz J., Zhang XM., Rott R. Comparison of hemag-glutinating, receptor-destroying, and acetylesterase activities of avirulent and virulent bovine coronavirus strains. Arch Virol. 1992. 125:193–204.

25). Zhang XM., Kousoulas KG., Storz J. Comparison of the nucleotide and deduced amino acid sequences of the S genes specified by virulent and avirulent strains of bovine coronaviruses. Virology. 1991. 183:397–404.

26). Rekik MR., Dea S. Comparative sequence analysis of a polymorphic region of the spike glycoprotein S1 subunit of enteric bovine coronavirus isolates. Arch Virol. 1994. 135:319–31.
27). Hasoksuz M., Sreevatsan S., Cho KO., Hoet AE., Saif LJ. Molecular analysis of the S1 subunit of the spike glycoprotein of respiratory and enteric bovine corona-virus isolates. Virus Res. 2002. 84:101–9.

28). Ko CK., Kang MI., Lim GK., Kim GY., Yoon SS., Park JT., Jeong C., Park SH., Park SJ., Kim YJ., Jeong JH., Kim SK., Park SI., Kim HH., Kim KY., Cho KO. Molecular characterization of HE, M, and E genes of winter dysentery bovine coronavirus circulated in Korea during 2002~2003. Virus Genes. 2006. 32:129–36.

29). Jeong JH., Kim GY., Yoon SS., Park SJ., Kim YJ., Sung CM., Shin SS., Lee BJ., Kang MI., Park NY., Koh HB., Cho KO. Molecular analysis of S gene of spike glycoprotein of winter dysentery bovine coronavirus circulated in Korea during 2002~2003. Virus Res. 2005. 108:207–12.

30). Park SJ., Jeong C., Yoon SS., Choy HE., Saif LJ., Park SH., Kim YJ., Jeong JH., Park SI., Kim HH., Lee BJ., Cho HS., Kim SK., Kang MI., Cho KO. Detection and characterization of bovine coronaviruses in fecal specimens of adult cattle with diarrhea during the warmer seasons. J Clin Microbiol. 2006. 44:3178–88.

31). Yang DK., Kweon CH., Kim BH., Park JK., So BJ., Song JY. Serological Survey of Bovine Coronavirus in Korea. J Bacteriol Virol. 2007. 37:105–9.

32). Han MG., Cheon DS., Zhang X., Saif LJ. Cross-protection against a human enteric coronavirus and a virulent bovine enteric coronavirus in gnotobiotic calves. J Virol. 2006. 80:12350–6.

33). Laude H., Talbot PJ., Levy GA. Corona related Virus: Functional Domains in the Spike Protein of Transmissible Gastroenteritis Virus. Plenum Press, New York. 1995. 299–394.
Go to : 
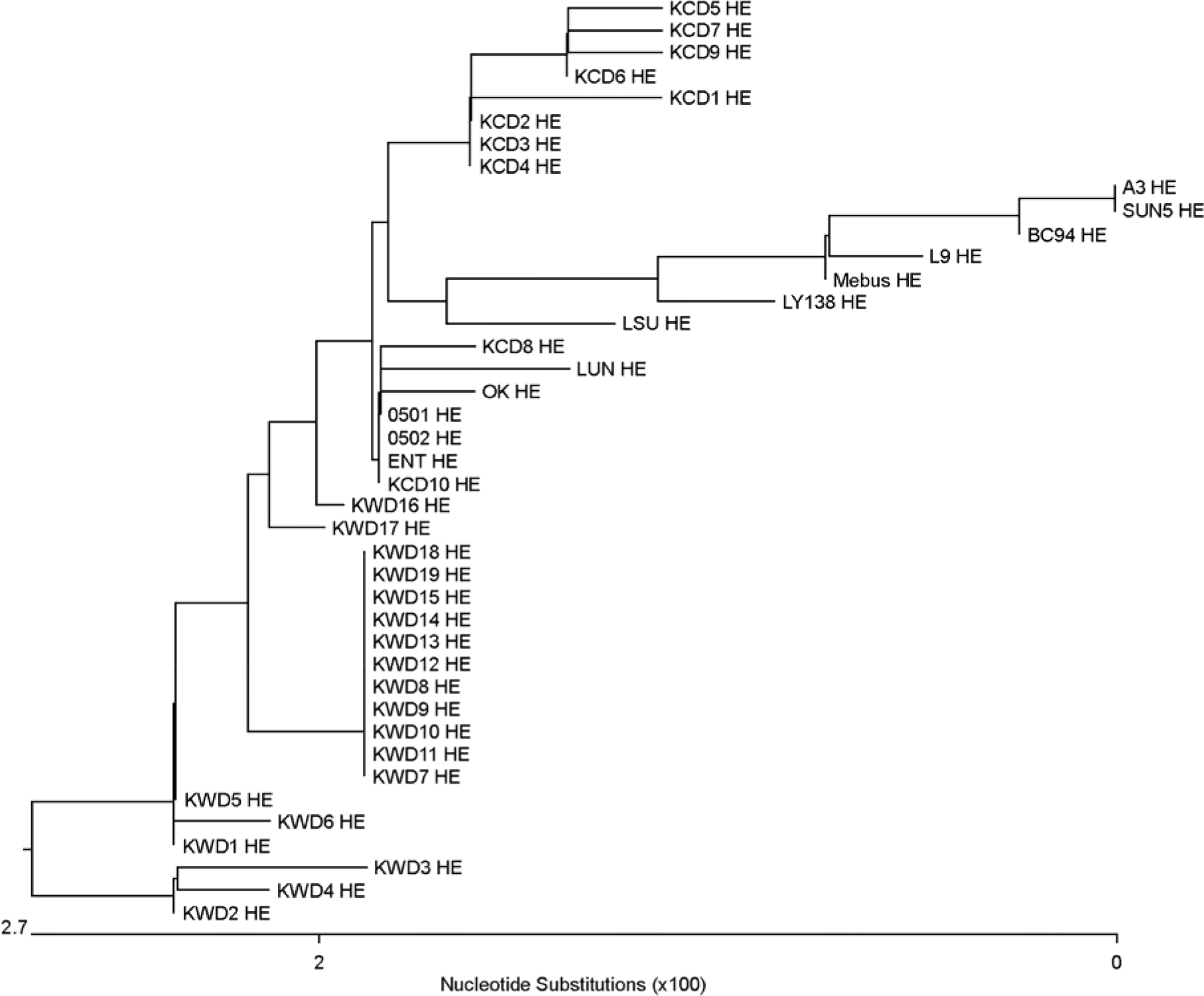 | Figure 1.Phylogenic tree of BCV HE. Mebus, L9, F15, LY-138, ENT, vaccine strain M64668, Korean vaccine BC94, SUN5, 0501, 0502, KWDs, and KCDs were analyzed by using Clustal W multiple alignment method of MegAlign program in DNAstar. |
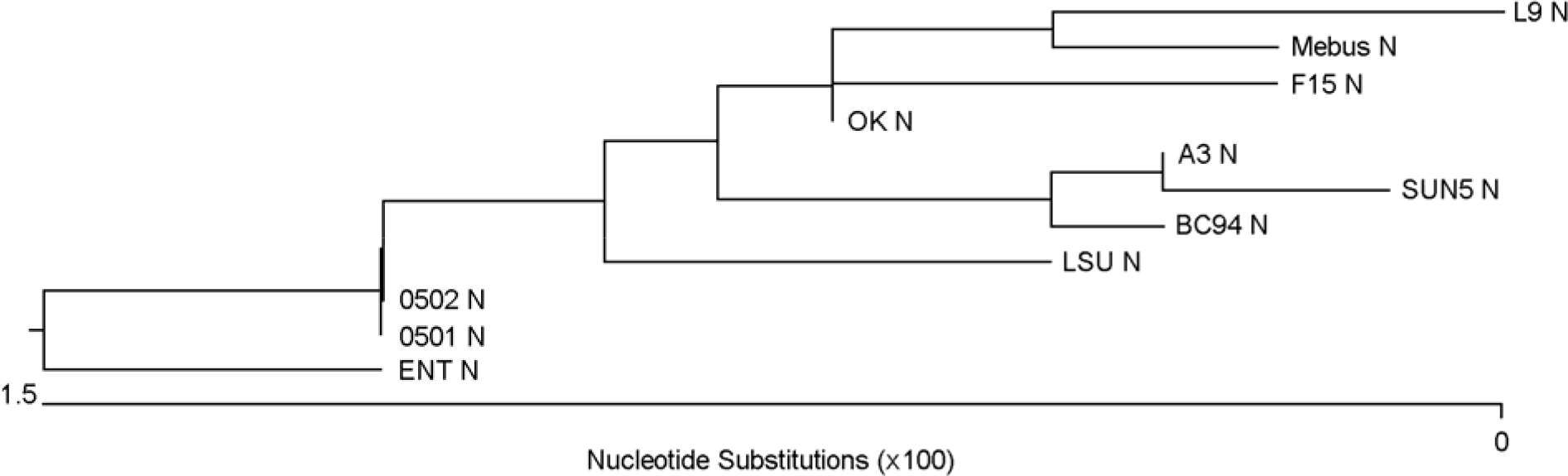 | Figure 2.Phylogenic tree of BCV N. Mebus, L9, F15, LY-138, ENT, vaccine strain M64668, Korean vaccine BC94, SUN5, 0501 and 0502 were analyzed by using Clustal W multiple alignment method of MegAlign program in DNAstar. |
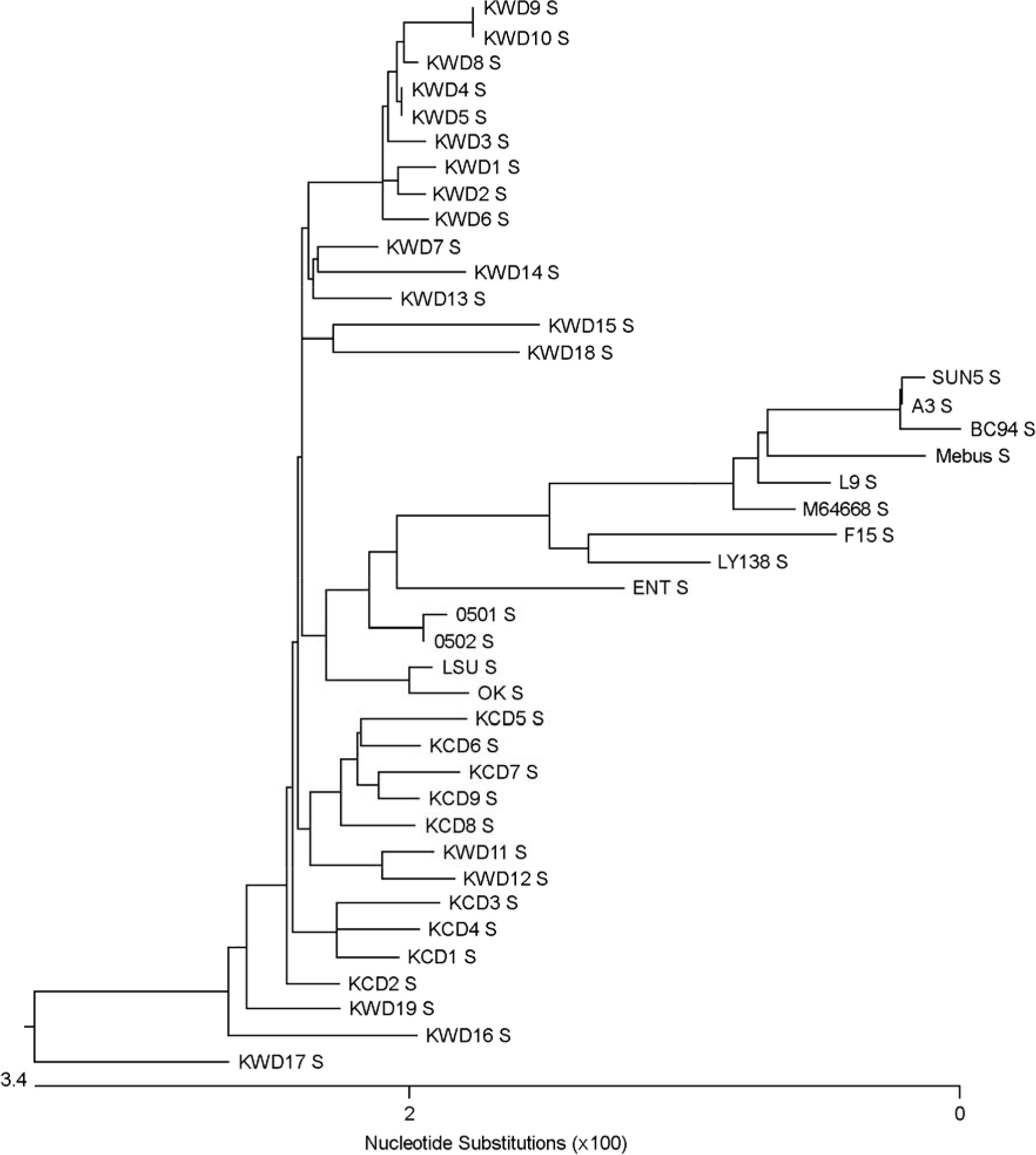 | Figure 3.Phylogenic tree of spike proteins of Mebus, L9, F15, LY-138, ENT, vaccine strain M64668, Korean vaccine BC94, SUN5, 0501, 0502, KWDs, and KCDs were made using Clustal W multiple alignment method of MegAlign program in DNAstar. |
 | Figure 4.Comparison of the deduced amino acid sequences of hypervariable region in spike proteins. Light-gray boxes contain respiratory bovine coronavirus (RBCV)-specific; dark-gray boxes contain virulent-specific and black boxes show significant sequence compared to recent Korean wide-spread BCoVs. |
Table 1.
The oligonucleotide primers of HE, N and S gene. Mebus strain (GenBank accession No. U00735) used for DNA cloning and sequencing
Table 2.
The GenBank accession numbers of reference strains of BCV vaccine and field isolates




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download