Abstract
Avian influenza (AI) virus infects both animal and human. Low pathogenic AI virus infections (some H7 and H9 subtypes) have been reported all over the world and pose a potential threat to the poultry industry. Vaccination is the most effective way to prevent virus infection. However, vaccination makes it difficult to differentiate between vaccinated chickens and infected chickens. In order to differentiate vaccinated chickens from naturally infected chickens, we adopted synthetic peptide-based enzyme-linked immunosorbent assay (ELISA) using the peptide sequences from nonstructural protein 1 (NS1) of H9N2. Five synthetic peptides were designed using Protein Variability Sever (http://imed.med.ucm.es/PVS/) and synthesized. NS1-1 ~ NS1-4 peptides failed to detect serum antibodies from both vaccinated and naturally infected chickens. NS1-5 peptide from the C-terminal NS1 protein detected serum antibody from naturally infected chickens but not vaccinated chickens. These results imply that NS1-5 peptide may be a useful tool to differentiate naturally infected chicken from vaccinated chicken as being used in the synthetic peptide-based ELISA.
Go to : 
REFERENCES
1). Yuen KY., Chan PK., Peiris M., Tsang DN., Que TL., Shortridge KF, et al. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998. 351:467–71.

3). Tumpey TM., Alvarez R., Swayne DE., Suarez DL. Diagnostic approach for differentiating infected from vaccinated poultry on the basis of antibodies to NS1, the nonstructural protein of influenza A virus. J Clin Microbiol. 2005. 43:676–83.

5). Dundon WG., Milani A., Cattoli G., Capua I. Progressive truncation of the Non-Structural 1 gene of H7N1 avian influenza viruses following extensive circulation in poultry. Virus Res. 2006. 119:171–6.

6). Webster RG., Peiris M., Chen H., Guan Y. H5N1 outbreaks and enzootic influenza. Emerg Infect Dis. 2006. 12:3–8.

7). Zhao S., Jin M., Li H., Tan Y., Wang G., Zhang R, et al. Detection of antibodies to the nonstructural protein (NS1) of avian influenza viruses allows distinction between vaccinated and infected chickens. Avian Dis. 2005. 49:488–93.

8). Banks J., Speidel EC., Harris PA., Alexander DJ. Phylogenetic analysis of influenza A viruses of H9 haemagglutinin subtype. Avian Pathol. 2000. 29:353–9.

9). Alexander DJ. Report on avian influenza in the Eastern Hemisphere during 1997~2002. Avian Dis. 2003. 47:792–7.

10). Guo YJ., Krauss S., Senne DA., Mo IP., Lo KS., Xiong XP, et al. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology. 2000. 267:279–88.

11). Lin YP., Shaw M., Gregory V., Cameron K., Lim W., Klimov A, et al. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci U S A. 2000. 97:9654–8.

12). Horimoto T., Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat Rev Microbiol. 2005. 3:591–600.

13). Lambrecht B., Steensels M., Van Borm S., Meulemans G., van den Berg T. Development of an M2e-specific enzyme-linked immunosorbent assay for differentiating infected from vaccinated animals. Avian Dis. 2007. 51:221–6.

14). Jeong OM., Kim MC., Kang HM., Ha GW., Oh JS., Yoo JE, et al. Validation of egg yolk antibody based C-ELISA for avian influenza surveillance in breeder duck. Vet Microbiol. 2010. 144:287–92.

15). Hale BG., Randall RE., Ortín J., Jackson D. The multifunctional NS1 protein of influenza A viruses. J Gen Virol. 2008. 89:2359–76.

16). Ghedin E., Sengamalay NA., Shumway M., Zaborsky J., Feldblyum T., Subbu V, et al. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature. 2005. 437:1162–6.

17). Wang W., Riedel K., Lynch P., Chien CY., Montelione GT., Krug RM. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA. 1999. 5:195–205.

18). Basler CF., Aguilar PV. Progress in identifying virulence determinants of the 1918 H1N1 and the Southeast Asian H5N1 influenza A viruses. Antiviral Res. 2008. 79:166–78.

19). Krug RM., Etkind PR. Cytoplasmic and nuclear virus-specific proteins in influenza virus-infected MDCK cells. Virology. 1973. 56:334–48.

Go to : 
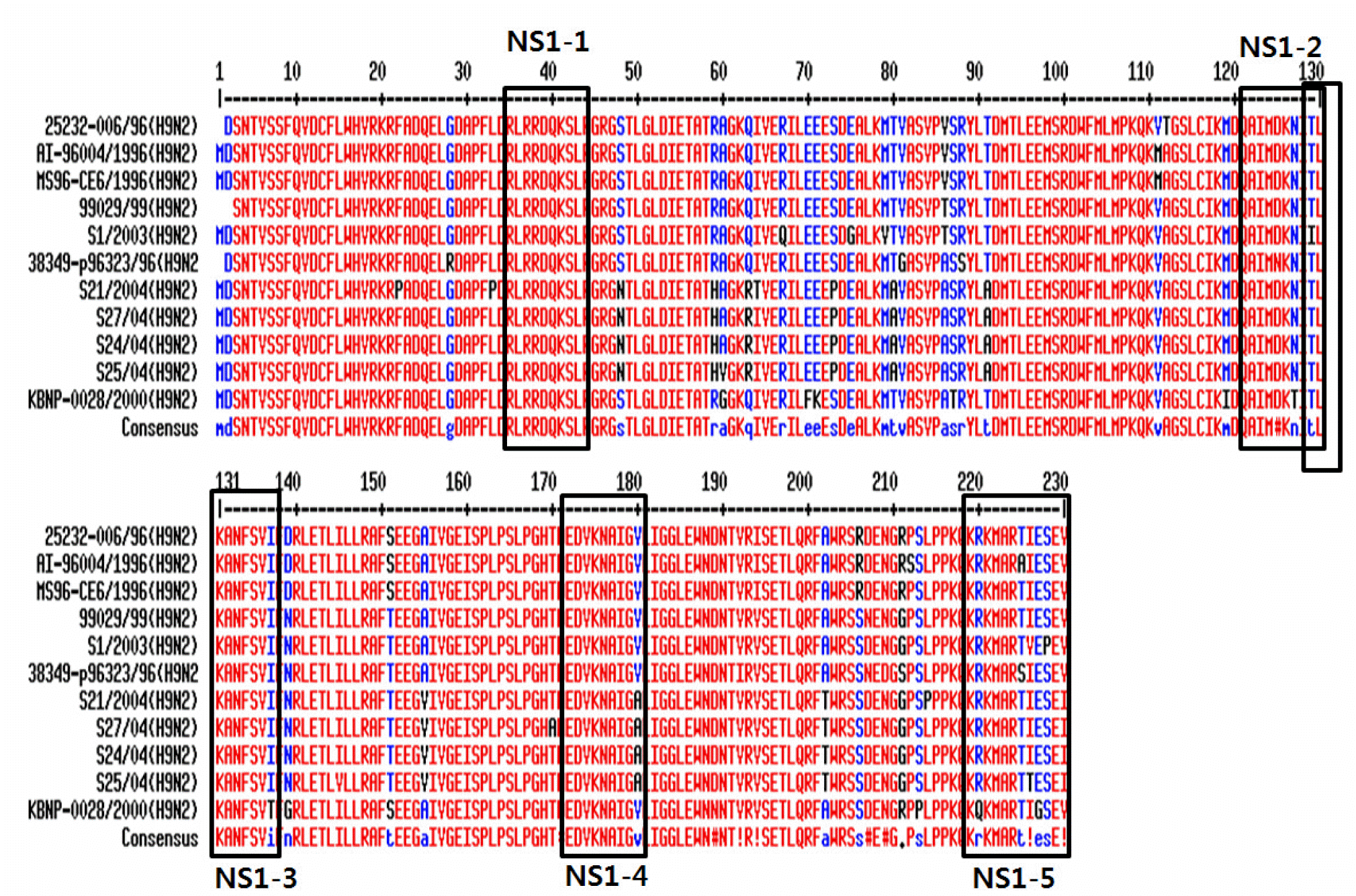 | Figure 1.Multiple sequence alignment of NS1 proteins from avian influenza virus reported in Korea using Multalin. The NS1 protein sequences were obtained from GenBank. Multiple sequence alignment analysis of NS1 proteins using Multalin displayed 84% homology. |
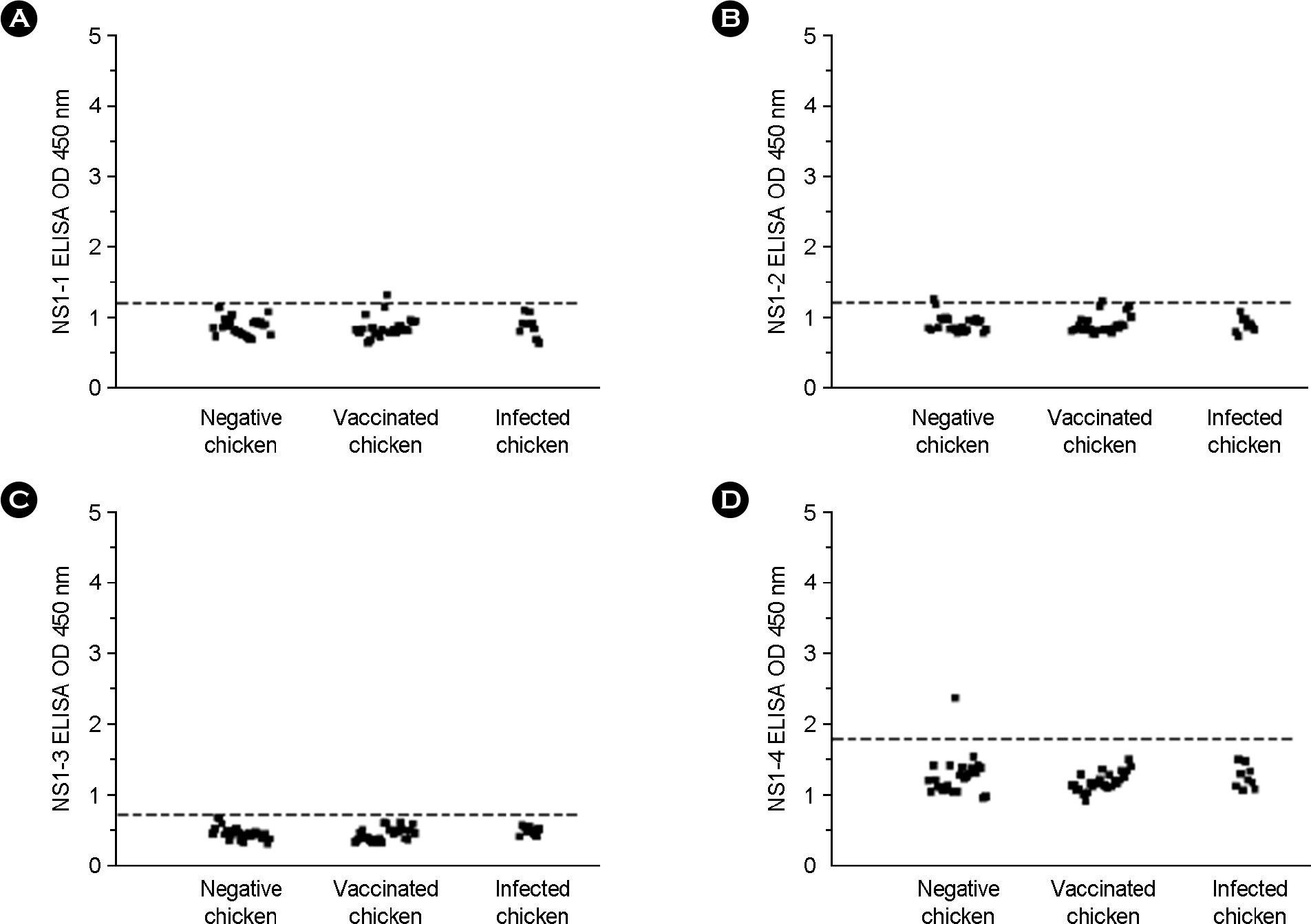 | Figure 2.Detection of anti-NS1 antibodies by synthetic peptide-based ELISA in chicken sera. (A) NS1-1, (B) NS1-2, (C) NS1-3, and (D) NS1-4. Dashed horizontal line represents the cut-off value calculated by adding an OD 450 nm mean value and two standard deviations (SD). The cut-off values for the test are (A) 1.122, (B) 1.127, (C) 0.601, and (D) 1.787. Serum sample is positive when OD value is greater than the line. |
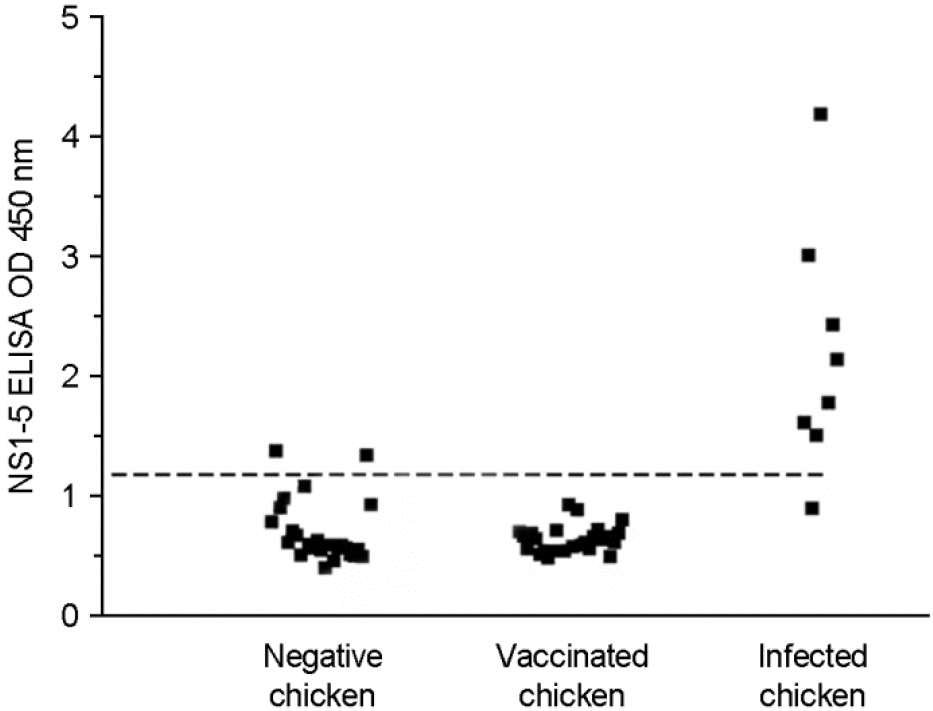 | Figure 3.Detection of anti-NS1 antibodies by synthetic NS1-5 peptide-based ELISA in chicken sera. Dashed horizontal line represents the cut-off value calculated by adding an OD 450 nm mean value and two standard deviations (SD). The cut-off value for the test is 1.257. Serum sample is positive when OD value is above line. |
Table 1.
GenBank accession number of NS1 proteins in Korea avian influenza




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download