Abstract
Foot-and-mouth disease (FMD) is an economically significant animal disease because of the speed of its transmission. Routine vaccination may not be effective for early protection in an outbreak situation. Small interfering RNA (siRNA) can be used as a rapid, effective, and an alternative antiviral approach. In this study, we screened 15 synthetic siRNAs to inhibit FMD virus replication in IBRS-2 cells and selected 10 siRNA sequences. Furthermore, we produced 7 adenoviruses expressing shRNA targeting conserved regions of FMDV, such as a leader sequence and nonstructural protein regions, and showed their antiviral effects. We compared the antiviral effects among them and compared between synthetic siRNAs and adenovirus-delivered siRNAs. In particular, the most efficient siRNA, 3C2, was the conserved sequence in the O, A, Asia 1, and C serotypes of FMDV and was located in the predicted loop structure. The pool of sequences including 3C2 and recombinant adenoviruses could be applied for multiple siRNAs and protection in a broad range of cells and animals.
Go to : 
REFERENCES
1). Pereira HG. Foot-and-mouth disease animals. New York, NY: Academic Press. 1981. 333–63.
3). Domingo E., Escarmís C., Baranowski E., Ruiz-Jarabo CM., Carrillo E., Núñez JI, et al. Evolution of foot-and-mouth disease virus. Virus Res. 2003. 91:47–63.

4). Knowles NJ., Samuel AR. Molecular epidemiology of foot-and-mouth disease virus. Virus Res. 2003. 91:65–80.

5). Gitlin L., Stone JK., Andino R. Poliovirus escape from RNA interference: short interfering RNA-target recognition and implications for therapeutic approaches. J Virol. 2005. 79:1027–35.

6). Saulnier A., Pelletier I., Labadie K., Colbère-Garapin F. Complete cure of persistent virus infections by antiviral iRNAs. Mol Ther. 2006. 13:142–50.
7). Jacque JM., Triques K., Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002. 418:435–8.

9). Hamasaki K., Nakao K., Matsumoto K., Ichikawa T., Ishikawa H., Eguchi K. Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett. 2003. 543:51–4.

10). Kahana R., Nakao K., Matsumoto K., Ichikawa T., Ishikawa H., Eguchi K. Inhibition of foot-and-mouth disease virus replication by small interfering RNA. J Gen Virol. 2004. 85:3213–7.

11). Chen W., Yan W., Du Q., Fei L., Liu M., Ni Z, et al. RNA interference targeting VP1 inhibits foot-and-mouth disease virus replication in BHK-21 cells and suckling mice. J Virol. 2004. 78:6900–7.

12). Chen W., Liu M., Jiao Y., Yan W., Wei X., Chen J, et al. Adenovirus-mediated RNA interference against foot-and-mouth disease virus infection both in vitro and in vivo. J Virol. 2006. 80:3559–66.
13). Souza AP., Haut L., Reyes-Sandoval A., Pinto AR. Recombinant viruses as vaccines against viral diseases. Braz J Med Biol Res. 2005. 38:509–22.

14). Reed LI., Muench H. A simple metheod of estimating fifty percent and points. Am J Hygiene. 1938. 27:493–7.
15). Roeder PL., Le Blanc Smith PM. Detection and typing of foot-and-mouth disease virus by enzyme-linked immunosorbent assay: a sensitive, rapid and reliable technique for primary diagnosis. Res Vet Sci. 1987. 43:225–32.

16). Thompson JD., Higgins DG., Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994. 22:4673–80.

17). Hall TA. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999. 41:95–8.
18). Root DE., Hacohen N., Hahn WC., Lander ES., Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006. 3:715–9.

19). Lo HL., Chang T., Yam P., Marcovecchio PM., Li S., Zaia JA, et al. Inhibition of HIV-1 replication with designed miRNAs expressed from RNA polymerase II promoters. Gene Ther. 2007. 14:1503–12.

20). Makinen PI., Koponen JK., Karkkainen AM., Malm TM., Pulkkinen KH., Koistinaho J, et al. Stable RNA interference: comparison of U6 and H1 promoters in endothelial cells and in mouse brain. J Gene Med. 2006. 8:433–41.
21). Schubert S., Grunweller A., Erdmann VA., Kurreck J. Local RNA target structure influences siRNA efficacy: systematic analysis of intentionally designed binding regions. J Mol Biol. 2005. 348:883–93.

22). de Almeida RS., Keita D., Libeau G., Albina E. Structure and sequence motifs of siRNA linked with in vitro down-regulation of morbillivirus gene expression. Antiviral Res. 2008. 79:37–48.
Go to : 
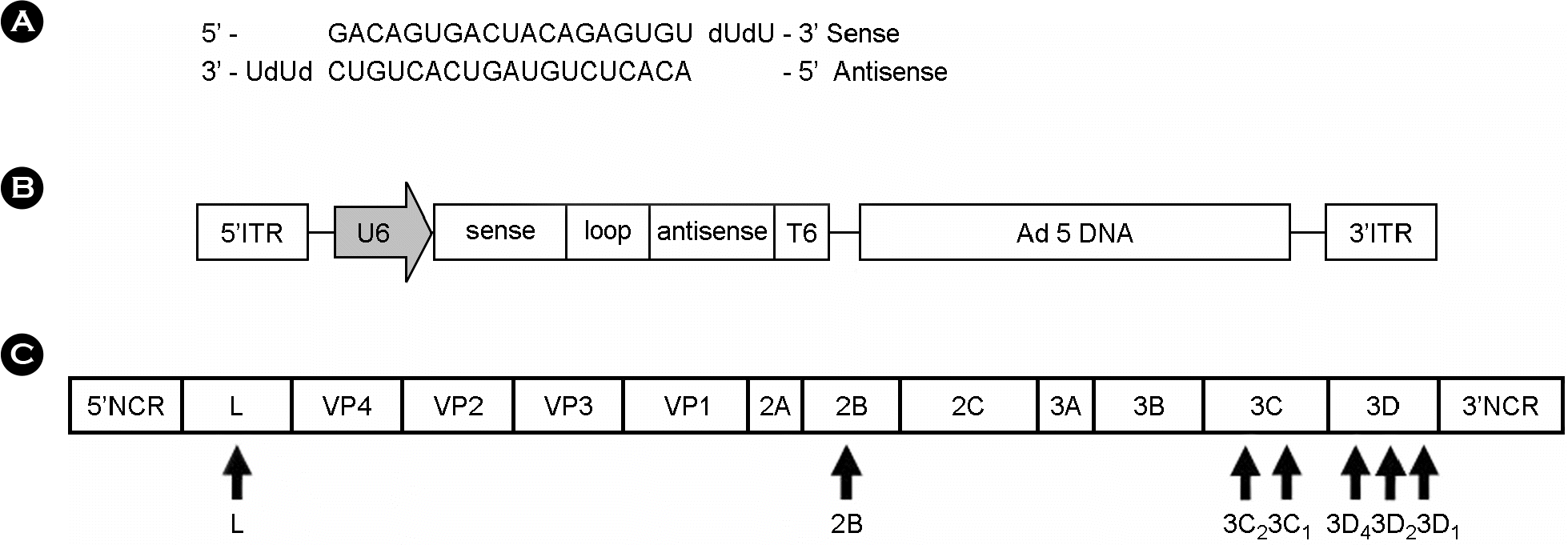 | Figure 1.Schematic representation of siRNA generation. (A) Schematic representation of annealed siRNA. (B) Schematic representation of recombinant adenovirus DNA. (C) Diagram of the FMDV full genome. The black arrows indicate the target region of shRNA. |
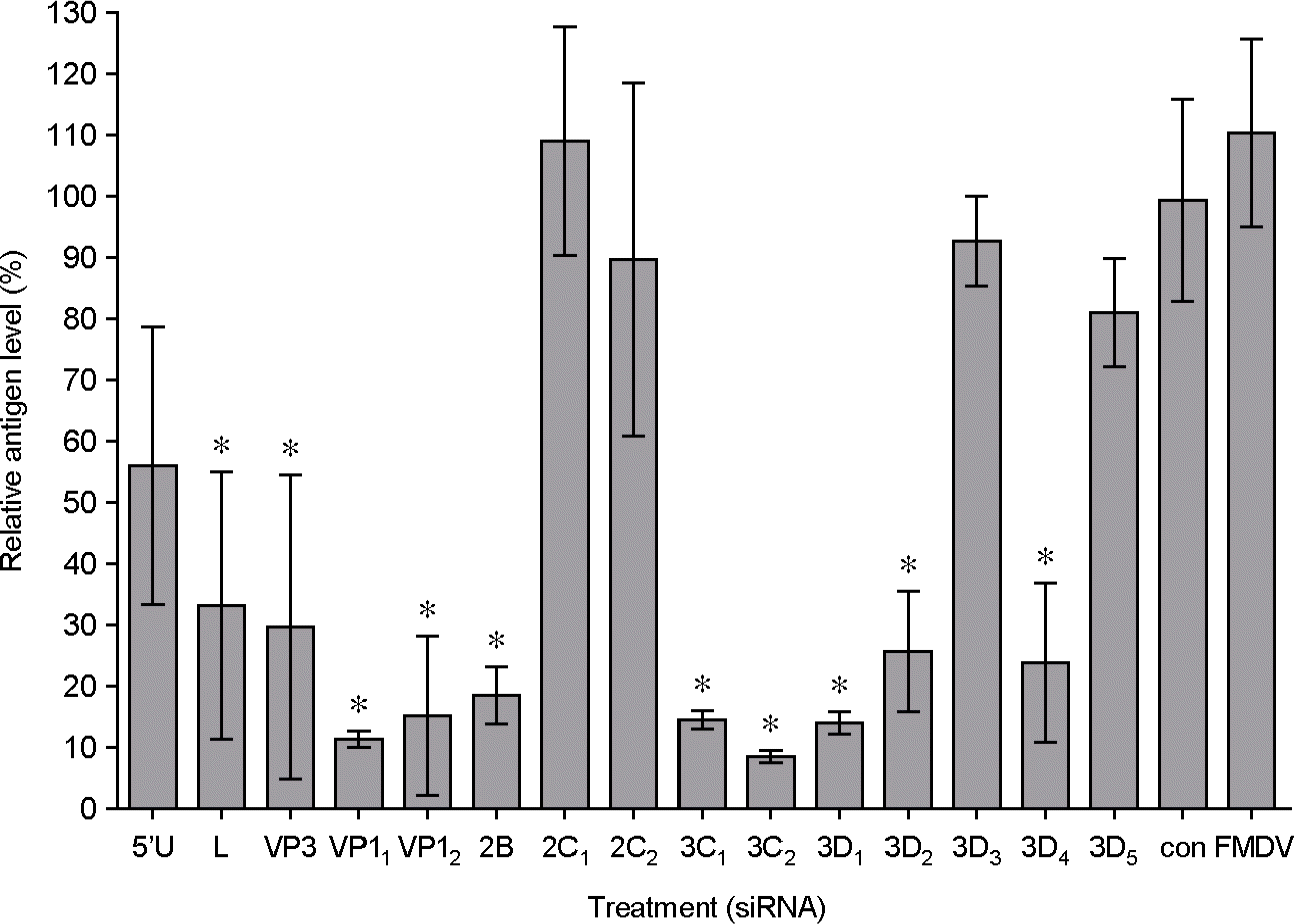 | Figure 2.Inhibition of FMDV replication in IBRS-2 cells by synthesized siRNA. IBRS-2 cells were transfected with 10 nM of each siRNA including negative control siRNA. After 12 h of transfection, the IBRS-2 cells were infected immediately with 100 TCID50 of FMDV (O/SKR/2002). The relative viral level was measured at 48 h p.i. using antigen ELISA (∗p < 0.05, t-test, a statistically significant differences between negative control (con) and others). Optical density (OD) was changed to relative antigen level (%) which was percentage of sample OD to negative control OD. Error bars indicate standard deviation (SD). |
 | Figure 3.Inhibition of FMDV replication in IBRS-2 cells by recombinant adenovirus. IBRS-2 cells were infected with AdshRNA at 3 × 106 TCID50. After 12 h of infection, the IBRS-2 cells were infected immediately with 100 TCID50 of FMDV (O/SKR/2002). The relative viral protein level measured at 48 h p.i. using antigen ELISA (∗p < 0.05 and ∗∗∗p < 0.0001, t-test, a statistically significant differences between control (Ad-Lamin A/C) and others. Optical density (OD) was changed to the relative antigen level (%), which was percentage of sample OD to negative control OD. Error bars indicate standard deviation (SD). |
 | Figure 4.Nucleotide sequence alignment of 3C2 siRNA region for seven serotypes of FMDV and RNA structure including 3C2 siRNA region. (A) Sequence alignment and editing were performed using CLUSTAL W (16) and BioEdit program (17). (B) RNA secondary structure of the partial 3C region including 3C2 siRNA sequences was obtained from the Mfold web server (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html). siRNA 3C2 sequences are included in the magnified figure and nucleotide T (thymine) represents U (uracil) in the RNA structure. Red lines represent G-C pairs and blue and green lines represent A-U and G-U pairs. |
 | Figure 5.Cross-inhibition of Ad-3C2 against O, A, and Asia 1 type of FMDV in IBRS-2 cells. IBRS-2 cells were infected with Ad-shRNA at 3 × 106 TCID50. After 12 h of infection, the IBRS-2 cells were infected immediately with 100 TCID50 of FMDV (O/SKR/2002, A22/IRQ, and Asia 1/MOG/05). The relative viral protein level measured at 24 and 48 h p.i. using antigen ELISA. Optical density (OD) was changed to the relative antigen level (%), which was percentage of sample OD to negative control OD. Error bars indicate standard deviation (SD). |
Table 1.
Target sequences of siRNAs used in this study




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download