Abstract
Echovirus 6 (ECV6) is the prevalent serotype detected in aseptic meningitis cases in Korea. To analyze the genetic variation of ECV6 isolates recently circulating in Korea, we determined the partial sequence of the VP1 capsid gene from 22 Korean ECV6 isolates and performed pairwise analysis against 42 reference strains from the GenBank database using MegAlign. The 22 Korean ECV6 isolates formed 3 distinct genetic clusters: Kor-lineage I, II, and III. The Korean ECV6 strains showed significant genetic diversity with 14.8~22.8% nucleotide divergence among the 3 different lineages. These ECV6 Kor-lineages were demonstrated to belong to different genetic clusters using VP1 sequence-based phylogenetic analysis, implying that the recently circulating Korean ECV6 strains have potential antigenic variation.
Go to : 
REFERENCES
1). King AMQ., Brown F., Christian P., Hovi T., Hyypiä T., Knowles NJ, et al. Picronaviridae. pp. p. 657–678. In. Virus taxonomy, Seventh Report of the International Committee on Taxonomy of Viruses. Van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB, editors. (Ed),. Academic Press;San Diego, Calif: 2000.
2). Stanway G., Brown F., Christian P., Hovi T., Hyypiä T., King AMQ, et al. Family Picornaviridae. pp. p. 757–778. In. Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. (Ed),. Elsevier Academic Press;London: 2005.
3). Ashwell MJ., Smith DW., Phillips PA., Rouse IL. Viral meningitis due to echovirus types 6 and 9: epidemiological data from Western Australia. Epidemiol Infect. 1996. 117:507–12.

4). Chomel JJ., Antona D., Thouvenot D., Lina B. Three ECHOvirus serotypes responsible for outbreak of aseptic meningitis in Rhône-Alpes region, France. Eur J Clin Microbiol Infect Dis. 2003. 22:191–3.

5). Abe O., Kimura H., Minakami H., Akami M., Inoue M., Saito A, et al. Outbreak of gastroenteritis caused by echovirus type 6 in an orphanage in Japan. J Infect. 2000. 41:285–6.

6). Boyd MT., Jordan SW., Davis LE. Fatal pneumonitis from congenital echovirus type 6 infection. Pediatr Infect Dis J. 1987. 6:1138–9.
7). Ventura KC., Hawkins H., Smith MB., Walker DH. Fatal neonatal echovirus 6 infection: autopsy case report and review of the literature. Mod Pathol. 2001. 14:85–90.

8). Khetsuriani N., Lamonte-Fowlkes A., Oberst S., Pallansch MA. Enterovirus surveillance–United States, 1970~2005; Centers for Disease Control and Prevention. MMWR Surveill Summ. 2006. 55:1–20.
9). Cabrerizo M., Echevarria JE., González I., de Miguel T., Trallero G. Molecular epidemiological study of HEV-B enteroviruses involved in the increase in meningitis cases occurred in Spain during 2006. J Med Virol. 2008. 80:1018–24.

10). Mirand A., Henquell C., Archimbaud C., Chambon M., Charbonne F., Peigue-Lafeuille H, et al. Prospective identification of enteroviruses involved in meningitis in 2006 through direct genotyping in cerebrospinal fluid. J Clin Microbiol. 2008. 46:87–96.

11). Richter J., Koptides D., Tryfonos C., Christodoulou C. Molecular typing of enteroviruses associated with viral meningitis in Cyprus, 2000~2002. J Med Microbiol. 2006. 55:1035–41.

12). Choi YJ., Park KS., Baek KA., Jung EH., Nam HS., Kim YB, et al. Molecular characterization of echovirus 30-associated outbreak of aseptic meningitis in Korea in 2008. J Microbiol Biotechnol. 2010. 20:643–9.
13). Jee YM., Cheon DS., Choi WY., Ahn JB., Kim KS., Chung YS, et al. Updates on enterovirus surveillance in Korea. Infect Chemother. 2004. 36:294–303.
14). Caggana M., Chan P., Ramsingh A. Identification of a single amino acid residue in the capsid protein VP1 of coxsackievirus B4 that determines the virulent phenotype. J Virol. 1993. 67:4797–803.

15). Dunn JJ., Chapman NM., Tracy S., Romero JR. Genomic determinants of cardiovirulence in coxsackievirus B3 clinical isolates: Localization to the 5′ nontranslated region. J Virol. 2000. 74:4787–94.

16). Knowlton KU., Jeon ES., Berkley N., Wessely R., Huber S. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J Virol. 1996. 70:7811–8.

17). Mateu MG. Antibody recognition of picornaviruses and escape from neutralization: a structural view. Virus Res. 1995. 38:1–24.

18). McPhee F., Zell R., Reimann BY., Hofschneider PH., Kandolf R. Characterization of the N-terminal part of the neutralizing antigenic site I of coxsackievirus B4 by mutation analysis of antigen chimeras. Virus Res. 1994. 34:139–51.

20). Oberste MS., Nix WA., Maher K., Pallansch MA. Improved molecular identification of enteroviruses by RT-PCR and amplicon sequencing. J Clin Virol. 2003. 26:375–7.

21). Mao N., Zhao L., Zhu Z., Chen X., Zhou S., Zhang Y, et al. An aseptic meningitis outbreak caused by echovirus 6 in Anhui province, China. J Med Virol. 2010. 82:441–5.

22). Papa A., Skoura L., Dumaidi K., Spiliopoulou A., Antoniadis A., Frantzidou F. Molecular epidemiology of echovirus 6 in Greece. Eur J Clin Microbiol Infect Dis. 2009. 28:683–7.

23). Nix WA., Oberste MS., Pallansch MA. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol. 2006. 44:2698–704.

24). Oberste MS., Maher K., Flemister MR., Marchetti G., Kilpatrick DR., Pallansch MA. Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. J Clin Microbiol. 2000. 38:1170–4.

25). Thompson JD., Higgins DG., Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994. 22:4673–80.

26). Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987. 4:406–25.
27). Baek K., Park K., Jung E., Chung E., Park J., Choi H, et al. Molecular and epidemiological characterization of enteroviruses isolated in Chungnam, Korea from 2005 to 2006. J Microbiol Biotechnol. 2009. 19:1055–64.

Go to : 
 | Figure 1.Gel electrophoresis of PCR products of VP1 gene of 22 ECV6 isolates from patients with aseptic meningitis; Lane M, 100 bp DNA ladder; Lane 1-22, 22 ECV6 isolates; Lane N, negative control; Lane P, positive control. |
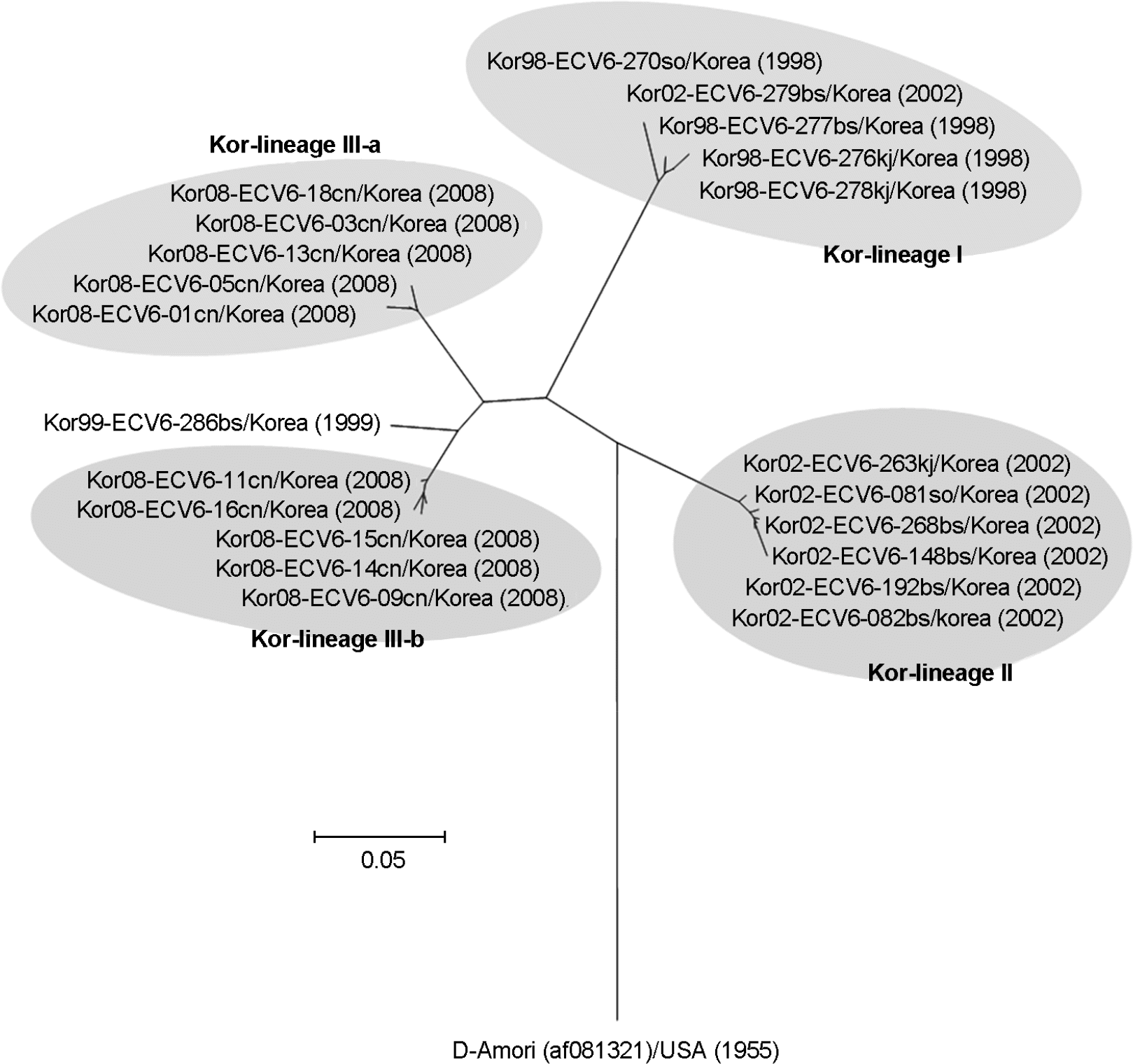 | Figure 2.Phylogenetic analysis based on a 286 bp sequence of the VP1 region of Korean ECV6 isolates. Nucleotide sequences were analyzed using the neighbor-joining method. |
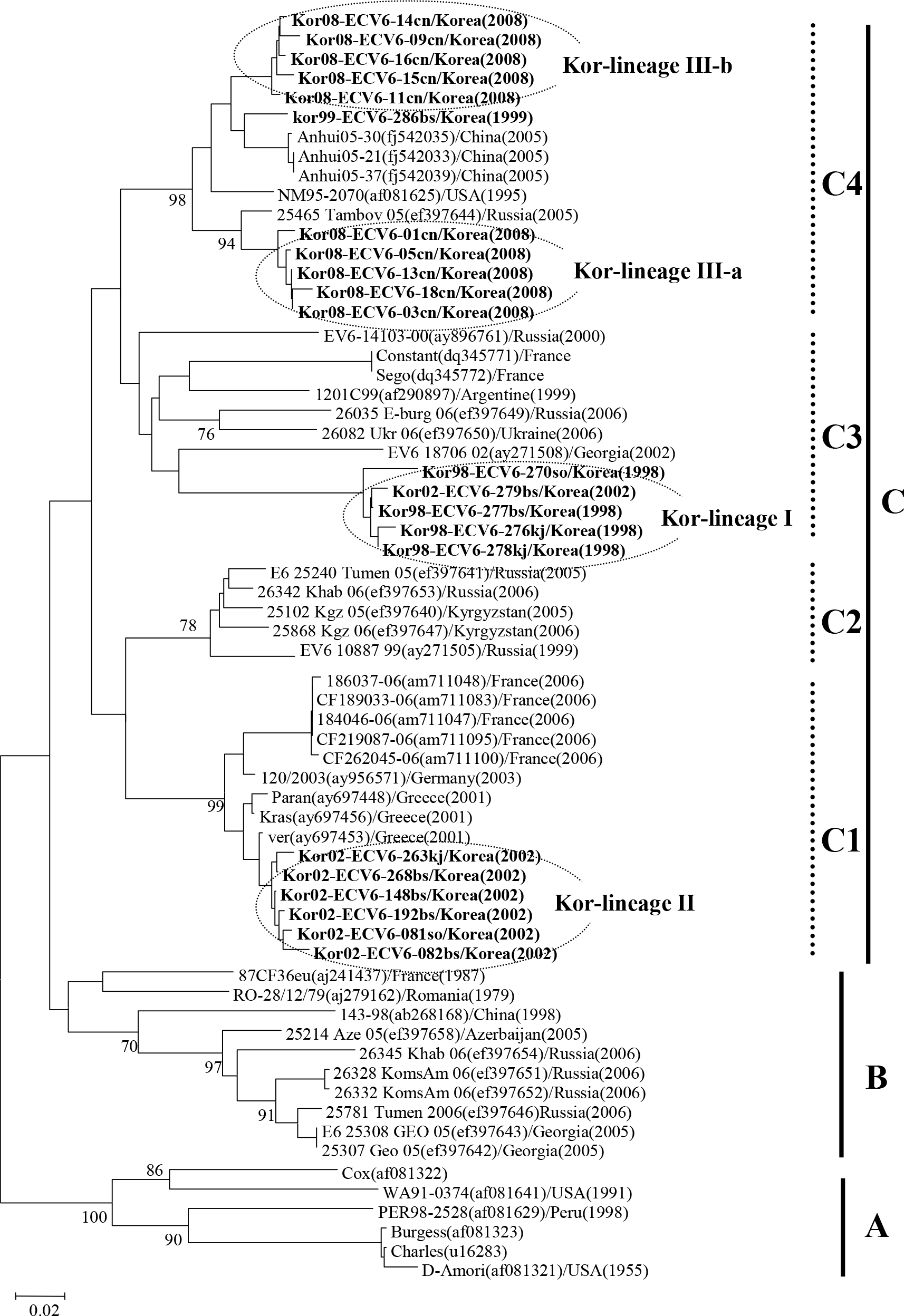 | Figure 3.Phylogenetic analysis based on a 286 bp sequence of the VP1 of ECV6 isolates. Nucleotide sequences were analyzed by the neighbor-joining method. The numbers at the branches indicate the bootstrap values for 1,000 replicates. |
Table 1.
Echovirus 6 strains isolated from patients' stool with aseptic meningitis
Table 2.
Percentage divergence of the nucleotide sequence of the VP1 coding region among the Korean lineage groups
| Kor-lineage I | Kor-lineage II | Kor-lineage III | D'Amori | |||
|---|---|---|---|---|---|---|
| a | b | |||||
| I | 14.0~18.9 | 17.3~18.7 | 15.3~17.7 | 24.2~24.5 | ||
| II | 13.1~16.5 | 14.5~17.3 | 21.0~22.2 | |||
| III | a | 7.2~9.8 | 23.8~25.2 | |||
| b | 23.1~24.5 | |||||
| D'Amori | ||||||
Table 3.
Percentage divergence of the nucleotide sequence of the VP1 coding region between the Korean lineage groups and cluster Cs




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download