Abstract
We have isolated 6 vancomycin resistant (VR) Enterococcus faecium and 5 VR-E. gallinarum. Vancomycin resistant enterococcus (VRE) isolates were resistant to multi-drugs, but susceptible to linezolid and quinupristin/dalfopristin. VRE isolates showed 10 VanA phenotypes and 1 VanB phenotype (E. gallinarum). However, all of them showed vanA genotype. vanA gene was detected on both genomic and plasmid DNA from all VRE isolates. Almost of VR-E. faecium had IS1216V which is worldwide type and almost of VR-E. gallinarum had IS1542 which is European type. IS1216V and IS1542 genes were not related with antibiotic types of VRE. Copy numbers of vanA were decreased in VRE with IS1216V or IS1542 but not in VRE with both ISs in broth without vancomycin. The copy numbers of vanA were significantly decreased in VanB phenotype of VRE with IS1542 in broth without vancomycin. Copy numbers of vanA were recovered in the presence of vancomycin. Growth time of reference E. faecium is faster than that of reference E. faecalis when cultured in the broth containing vancomycin. Reference strains cultured in the broth containing vancomycin showed intermediate resistance or resistance to antibiotics without acquisition of van genes. Naturally, multidrug-resistant E. faecium might be fast adapted in the presence of vancomycin compared to E. faecalis. Taken together, VanA phenotype E. gallinarum as well as E. feacium have been increasing in nosocomial infection and showed acquired inducible resistance. E. faecium and E. faecalis showed intermediate resistance in long exposure of vancomycin without acquisition of vanA.
Go to : 
REFERENCES
1). Sood S., Malhotra M., Das BK., Kapil A. Enterococcal infections & antimicrobial resistance. Indian J Med Res. 2008. 128:111–21.
2). Schaberg DR., Culver DH., Gaynes RP. Major trends in the microbial etiology of nosocomial infection. Am J Med. 1991. 91:72S–5S.

3). Livornese LL Jr., Drus S., Samel C. Hospital-acquired infection with vancomycin-resistant Enterococcus faecium transmitted by electronic thermometers. Ann Intern Med. 1992. 117:112–6.
4). Murray BE., Singh KV., Lopardo HA., Patterson JE., Zervos MJ, et al. Evidence for clonal spread of a single strain of β-lactamase-producing Enterococcus faecalis to six hospitals in five states. J Infect Dis. 1991. 163:780–5.
5). Murray BE., Lopardo HA., Rubeglio EA., Frosolono M., Singh KV. Intrahospital spread of single gentamicin-resistant, β-lactamase-producing strain of Enterococcus faecalis in Argentina. Antimicrob Agents Chemother. 1992. 36:230–2.
6). Porwancher R., Sheth A., Remphrey S., Taylor E., Hinkle C., Zervos M. Epidemiological study of hospital-acquired infection with vancomycin-resistant Enterococcus faecium: possible transmission by an electronic ear-probe thermometer. Infect Control Hosp Epidemiol. 1997. 18:771–3.
7). Leclercq R., Derlot E., Duval J., Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988. 319:157–61.
8). Arthur M., Courvalin P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 1993. 37:1563–71.

9). Leclercq R., Derlot E., Weber M., Duval J., Courvalin P. Transferable vancomycin and teicoplanin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1989. 33:10–5.
10). Noble WC., Virani Z., Cree RGA. Co-transfer of vancomycin and other resistance gens from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol Lett. 1992. 72:195–8.
11). Brown AR., Townsley AC., Amyes SG. Diversity of Tn1546 elements in clinical isolates of glycopeptide-resistant enterccocci from Scottish hospitals. Antimicrob Agents Chemother. 2001. 45:1309–11.
12). Darini AL., Palepou MF., Woodford N. Nucleotide sequence of IS1542, an insertion sequence identified within VanA glycopeptide resistance elements of enterococci. FEMS Microbiol Lett. 1999. 173:341–6.
13). Handwerger S., Skoble J., Discotto LF., Pucci MJ. Heterogeneity of the vanA gene cluster in clinical isolates of enterococci from the Northeastern United States. Antimicrob Agents Chemother. 1995. 39:362–8.
14). Huh JY., Lee WG., Lee K., Shin WS., Yoo JH. Distribution of insertion sequences associated with Tn1546-like elements among Enterococcus faecium isolates from patients in Korea. J Clin Microbiol. 2004. 42:1897–902.
15). Jensen LB., Ahrens P., Dons L., Jones RN., Hammerum AM., Aarestrup FM. Molecular analysis of Tn1546 in Enterococcus faecium isolated from animals & humans. J Clin Microbiol. 1998. 36:437–42.
16). Simonsen GS., Myhre MR., Dahl KH., Olsvik O., Sundsfjord A. Type ability of Tn1546-like elements in vancomycin-resistant enterococci using long-range PCRs and specific analysis of polymorphic regions. Microb Drug Resist. 2000. 6:49–57.
17). Arthur M., Molinas C., Courvallin P. The VanS VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1992. 174:2582–91.
18). Hashimoto Y., Tanimoto K., Ozawa Y., Murata T., Ike Y. Amino acid substitutions in the VanS sensor of the VanA type vancomycin resistant enterococcus strains result in high-level vancomycin resistance and low-level teicoplanin resistance. FEMS Microbiol Lett. 2000. 185:247–54.
19). Lee WG., Huh JY., Cho SR., Lim YA. Reduction in glycopeptide resistance in vancomycin-resistant enterococci as a result of vanA cluster rearrangements. Antimicrob Agents Chemother. 2004. 48:1379–81.
20). Arias CA., Martín-Martinez M., Blundell TL., Arthur M., Courvalin P., Reynolds PE. Characterization and modelling of VanT: a novel, membrane-bound, serine racemase from vancomycin-resistant Enterococcus gallinarum BM4174. Mol Microbiol. 1999. 31:1653–64.
21). Park IJ., Lee WG., Shin JH., Lee KW., Woo GJ. VanB phenotype-vanA genotype Enterococcus faecium with heterogeneous expression of teicoplanin resistance. J Clin Microbiol. 2008. 46:3091–3.
22). Henrique PM., Palazzo IC., Zanella RC., Darini AL. Molecular characterization of enterococci harboring genotype and phenotype incongruence related to glycopeptide resistance isolated in Brazilian hospitals. Mem Inst Oswaldo Cruz. 2008. 103:301–5.

23). Baptista M., Depardieu F., Courvalin P., Arthur M. Specificity of induction of glycopeptide resistance genes in Enterococcus faecalis. Antimicrob Agents Chemother. 1996. 40:2291–5.
24). Loeb M., Salama S., Armstrong-Evans M., Capretta G., Olde J. A case-control study to detect modifiable risk factors for colonization with vancomycin-resistant enterococci. Infect Control Hosp Epidemiol. 1999. 20:760–3.

25). Ratanasuwan W., Iwen PC., Hinrichs SH., Rupp ME. Bacteremia due to motile Enterococcus species: clinical features and outcomes. Clin Infect Dis. 1999. 28:1175–7.
Go to : 
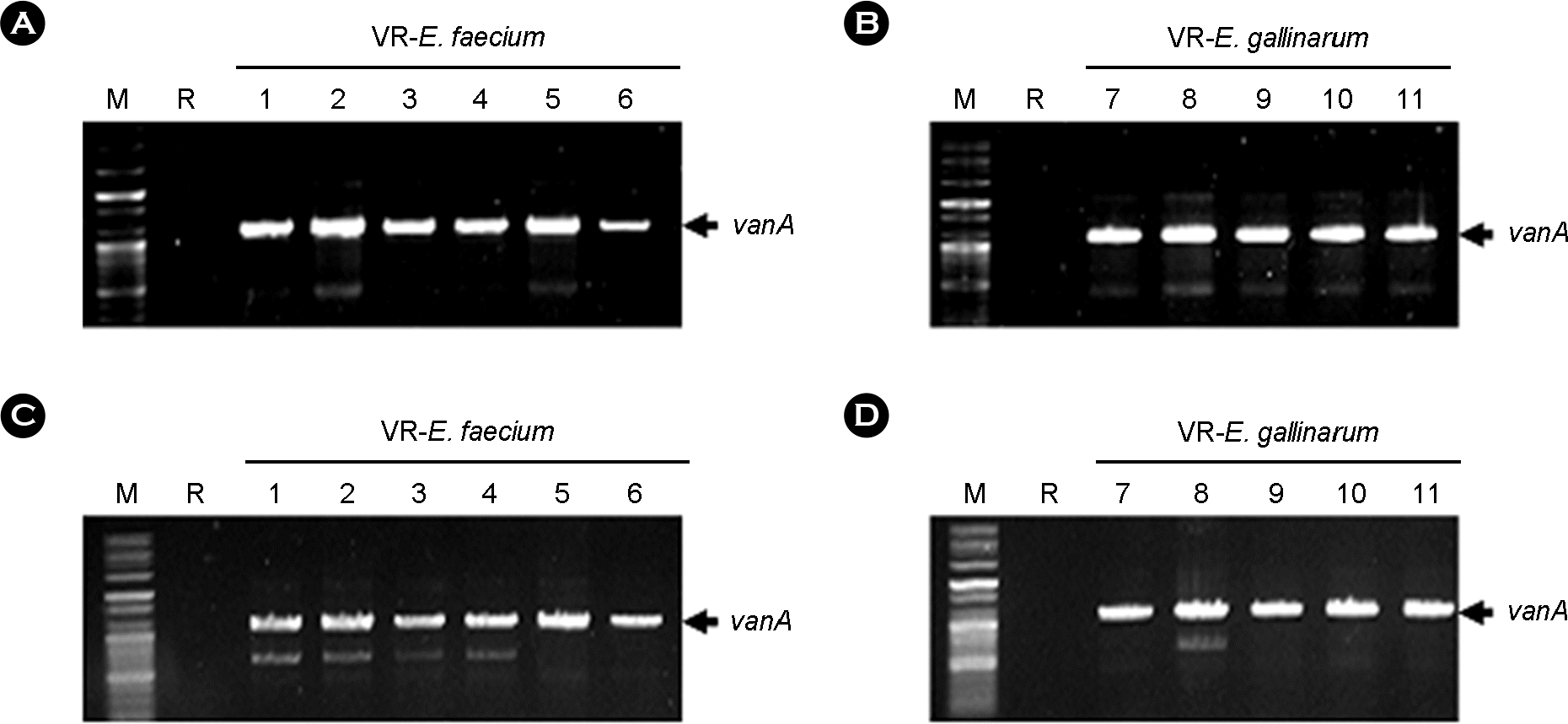 | Figure 1.Detection of vanA gene in genomic and plasmid DNA of VRE isolates from patients by PCR using vanA primer set. Amplified PCR products were analyzed by 1% agarose gel electrophoresis. A, B: Genomic DNA, C, D: plasmid DNA. vanA was detected in both genomic and plasmid DNA from all of VRE isolates except for reference Enterococcus faecium (R; ATCC 19434). M: 100 bp plus ladder DNA molecular size marker, R: reference Enterococcus faecium (ATCC 19434). |
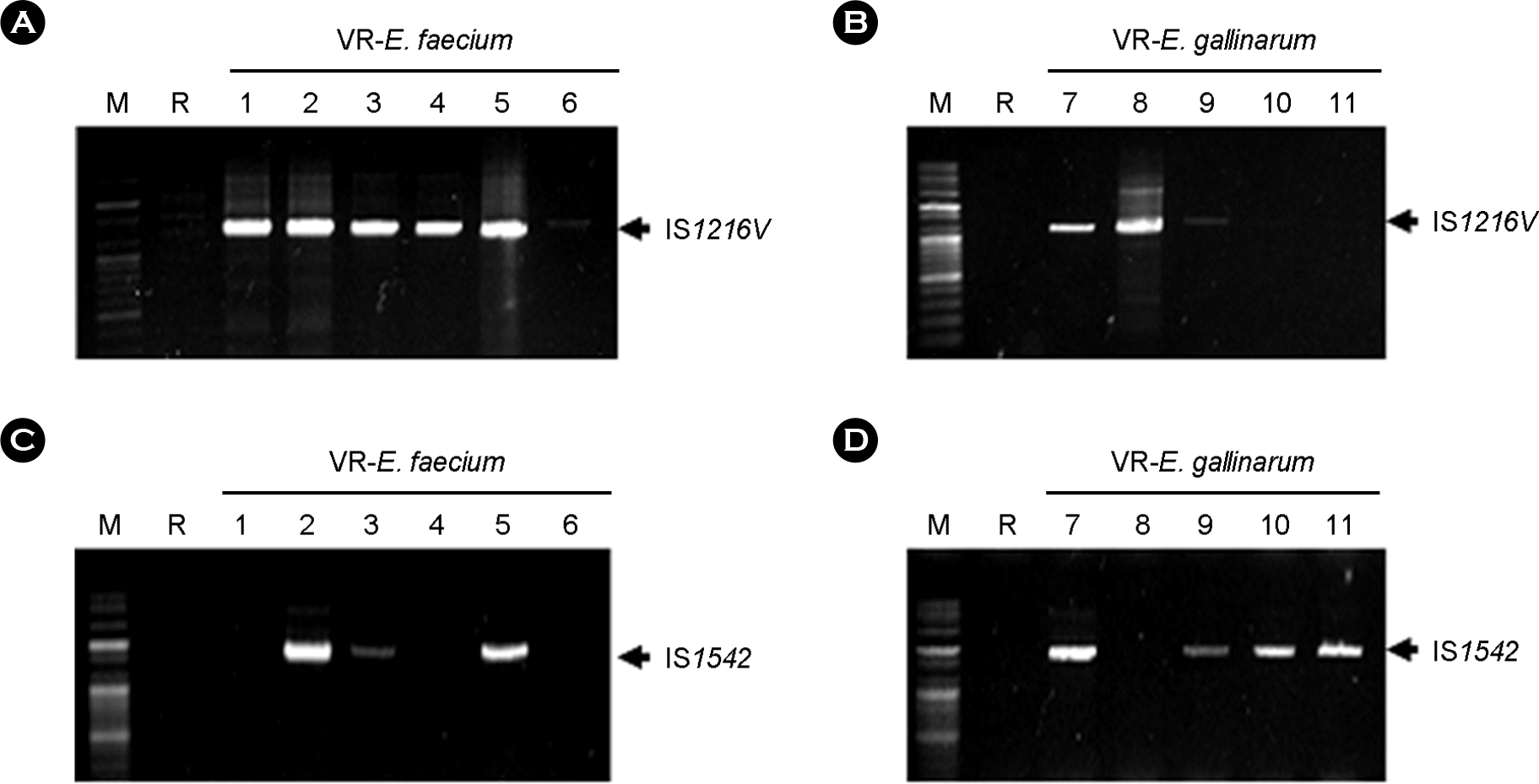 | Figure 2.Detection of IS1216V and IS1542 gene in plasmid DNA of VRE isolates from patients by PCR using specific primer set. The amplified PCR products were analyzed by 1% agarose gel electrophoresis. Plasmid DNA was extracted by using LaboPass Plasmid Mini Purification kit. IS1216V was detected strongly in plasmids from VRE1-5 and 7-8 except for reference Enterococcus faeciem (ATCC 19434), 6, 10-11. The gene was weakly amplified in those from VRE9. IS1542 was detected in VRE2, 3, 5, 7 and 9-11. M: 100 bp plus ladder DNA molecular size marker, R: reference Enterococcus faecium (ATCC 19434), VRE, vancomycin-resistant enterococci. |
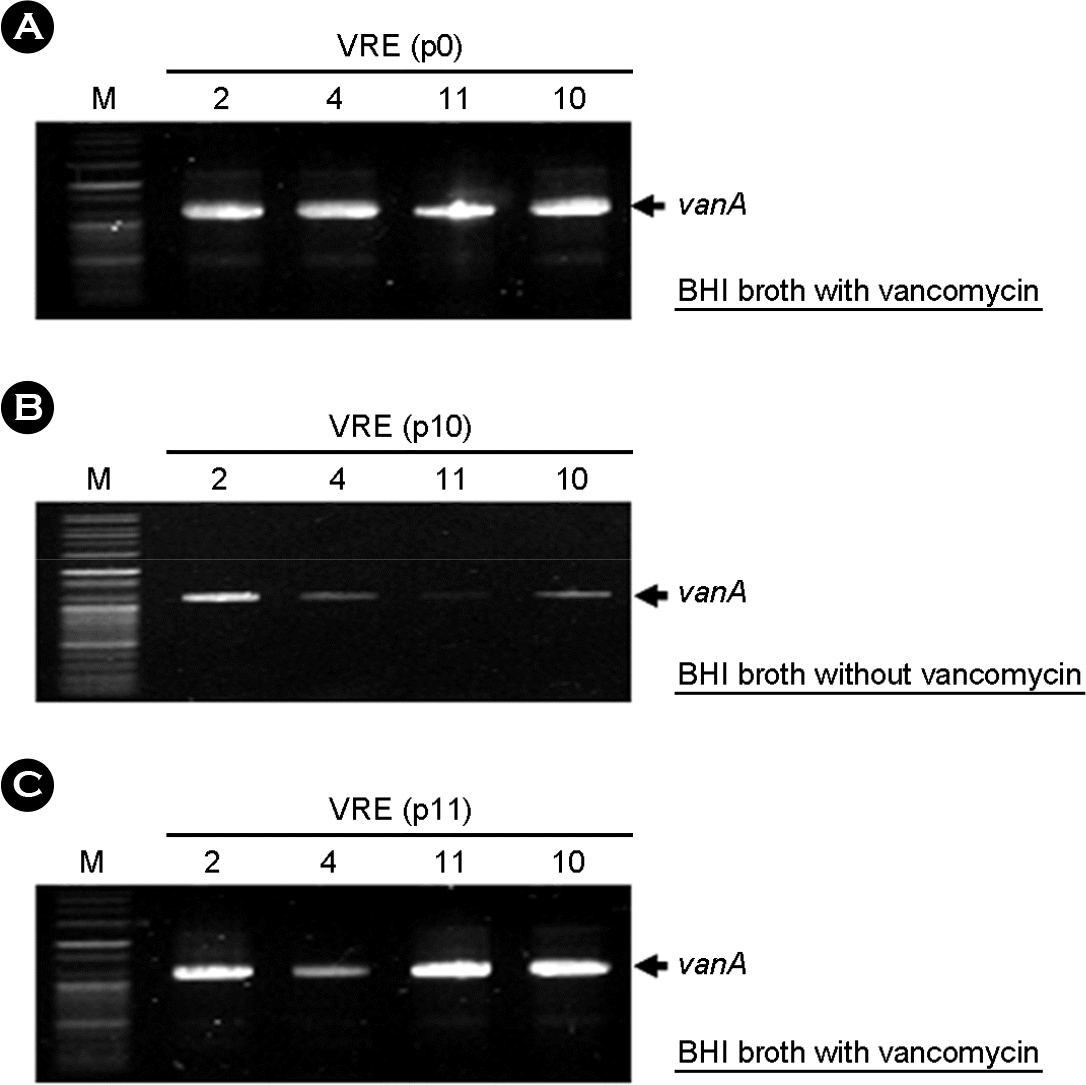 | Figure 3.Change of vanA gene in plasmid of VRE isolates through serial passage using BHI broth without vancomycin. vanA was detected weakly in plasmids from VRE 7 and 9, which are E. gallinarum isolates, but not changed in those from VRE 4 and 11, Enterococcus faecium isolates. VRE 2 & 4: VR-E. faecium, VRE10 & 11: VR-E. gallinarum, VRE2: vanA phenotype with IS1216V and IS1542, VRE4: vanA phenotype with only IS1216V, VRE10: vanA phenotype with only IS1542, VRE11: vanB phenotype with only IS1542. M: 100 bp plus ladder DNA molecular size marker, VRE, vancomycin-resistant enterococci. p10: 10th passaged VRE in normal growth media. p11: changed media to broth with 10 μg/ml of vancomycin |
Table 1.
Sequences of primers used for determination of genotypes of vancomycin-resistant enterococci
Table 2.
Difference of antimicrobial susceptibility among six isolated VR-E. faecium
Table 3.
Difference of antimicrobial susceptibility among five isolated VR-E. gallinarum
Table 4.
Changes of growth time of reference E. faecalis and E. faecium in BHI broth with different concentration of vancomycin
| Enterococci | Concentration of vancomycin (μg/ml) | Passage number | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 16 | 20 | ||
| E. faecalis | 4 | 72a | 72 | 48 | 48 | 24 | 24 | 24 | 24 |
| E. faecium | 4 | 144 | 48 | 24 | 24 | 24 | 24 | 24 | 24 |
| E. faecalis | 10 | 240 | 360 | 144 | 72 | 72 | 72 | 48 | 48 |
| E. faecium | 10 | 312 | 96 | 96 | 96 | 72 | 72 | 48 | 48 |
Table 5.
Results by GPI, GPS-451 and disk diffusion test to final passaged reference E. faecalis and E. faecium in broth with 4 and 10 μg/ml of vancomycin




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download