Abstract
As part of an initial inquiry into the function of the outer membrane proteins (OMPs) of Helicobacter pylori Korean strain 51, we have conducted an extensive proteome analysis via quadrupole time of flight (Q-TOF) mass spectrometry (MS). Fifty one OMPs of H. pylori were purified using sarcosine and resolved via two-dimensional electrophoresis with immobilized pH gradient strips. The most abundant proteins were observed in the alkaline pI regions (6.0~11.0) at molecular masses between 10~100 KDa. Here, 15 spots were identified, representing 9 types of genes (KHP0852, KHP0853, KHP1353, KHP1017, KHP0172, KHP0076, KHP0617, KHP1069, KHP0614) from the sarcosin-insoluble fraction of H. pylori 51. These may be employed in the characterization of the OMPs of H. pylori 51, which will help to identify new potential target proteins for vaccine development and drug therapy.
Go to : 
REFERENCES
1). Tomb JF., White O., Kerlavage AR., Clayton RA., Sutton GG., Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997. 388:539–47.
2). Matsuhisa TM., Yamada NY., Kato SK., Matsukura NM. Helicobacter pylori infection, mucosal atrophy and intestinal metaplasia in Asian populations: a comparative study in age-, gender- and endoscopic diagnosis-matched subjects. Helicobacter. 2003. 8:29–35.
3). Youn HS., Baik SC., Cho YK., Woo HO., Ahn YO., Kim K, et al. Comparison of Helicobacter pylori Infection between Fukuoka, Japan and Chinju, Korea. Helicobacter. 1998. 3:9–14.
4). Zhou W., Yamazaki S., Yamakawa A., Ohtani M., Ito Y., Keida Y, et al. The diversity of vacA and cagA genes of Helicobacter pylori in East Asia. FEMS Immunol Med Microbiol. 2004. 40:81–7.
5). Marcelli SW., Chang HT., Chapman T., Chalk PA., Miles RJ., Poole RK. The respiratory chain of Helicobacter pylori: identification of cytochromes and the effects of oxygen on cytochrome and menaquinone levels. FEMS Microbiol Lett. 1996. 138:59–64.
6). Yamaoka Y., Ojo O., Fujimoto S., Odenbreit S., Haas R., Gutierrez O, et al. Helicobacter pylori outer membrane proteins and gastroduodenal disease. Gut. 2006. 55:775–81.
7). Cho MJ., Jeon BS., Park JW., Jung TS., Song JY., Lee WK, et al. Identifying the major proteome components of Helicobacter pylori strain 26695. Electrophoresis. 2002. 23:1161–73.
8). Drouet EB., Denoyel GA., Boude M., Wallano E., Andujar M., de Demontclos HP. Characterization of an immuno-reactive species-specific 19-kilodalton outer membrane protein from Helicobacter pylori by using a monoclonal antibody. J Clin Microbiol. 1991. 29:1620–4.
9). Illingworth DS., Walter KS., Griffiths PL., Barclay R. Siderophore production and iron-regulated envelope proteins of Helicobacter pylori. Zentralbl Baketeriol. 1993. 280:113.
10). Heukeshoven J., Dernick R. Improved silver staining procedure for fast staining in Phast System Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis. 1988. 9:28–32.
11). Guerreiro N., Redmond JW., Rolfe BG. Djordjevic MA. New Rhizobium legaminosarum flavonoid-induced proteins revealed by proteome analysis of differentially displayed proteins. Mol Plant Microbe Interact. 1997. 10:506–16.
12). O'Connell KL., Stalts JT. Identification of mouse liver proteins on two-dimensional electrophoresis gels by matrix-assisted larser desorption/ionization mass spectrometry of in situ enzymatic digests. Electrophoresis. 1997. 18:349–59.
13). Jensen ON., Larsen MR., Roepstorff P. Mass spectrometric identification and microcharacterization of proteins from electrophoretic gels: strategies and applications. Proteins. 1988. ;Suppl. 2:74–89.

14). Langen H., Gray C., Röder D., Juranville JF., Takacs B., Fountoulakis M. From genome to proteome: protein map of Haemophilus influenzae. Electrophoresis. 1997. 18:1184–92.
15). Alm RA., Ling LS., Moir DT., King BL., Brown ED., Doig PC, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999. 397:176–80.
16). Papac DI., Shahrokh Z. Mass spectrometry innovations in drug discovery and development. Pharm Res. 2001. 18:131–45.
17). Yoon SH., Kim MS. Development of a time-resolved method for photodissociation mechanistic study of protonated peptides: use of a voltage-floated cell in a tandem time-of-flight mass spectrometer. J Am Soc Mass Spectrom. 2007. 18:1729–39.

18). Baik SC., Kim KM., Song SM., Kim DS., Jun JS., Lee SG, et al. Proteomic Analysis of sarcosine-insoluble outer membrane fraction of Helicobacter pylori strain 26695. J Bacteriol. 2004. 186:949–55.
19). Antelmann H., Scharf C., Hecker M. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J Bacteriol. 2000. 182:4478–90.
20). Xu Q., Peek RM Jr., Miller GG., Blaser MJ. The Helicobacter pylori genome is modified at CATG by the product of hpyIM. J Bacteriol. 1997. 179:6807–15.
21). Ando T., Xu Q., Torres M., Kusugami K., Israel DA., Blaser MJ. Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol Microbiol. 2000. 37:1052–65.
22). McGovern KJ., Blanchard TG., Gutierrez JA., Czinn SJ., Krakowka S., Youngman P. γ-glutamyltransferase is a Helicobacter pylori virulence factor but is not essential for colonization. Infect Immun. 2001. 69:4168–73.
23). Bumann D., Aksu S., Wendland M., Janek K., Zimny-Arndt U., Sabarth N, et al. Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori. Infect Immun. 2002. 70:3396–403.
Go to : 
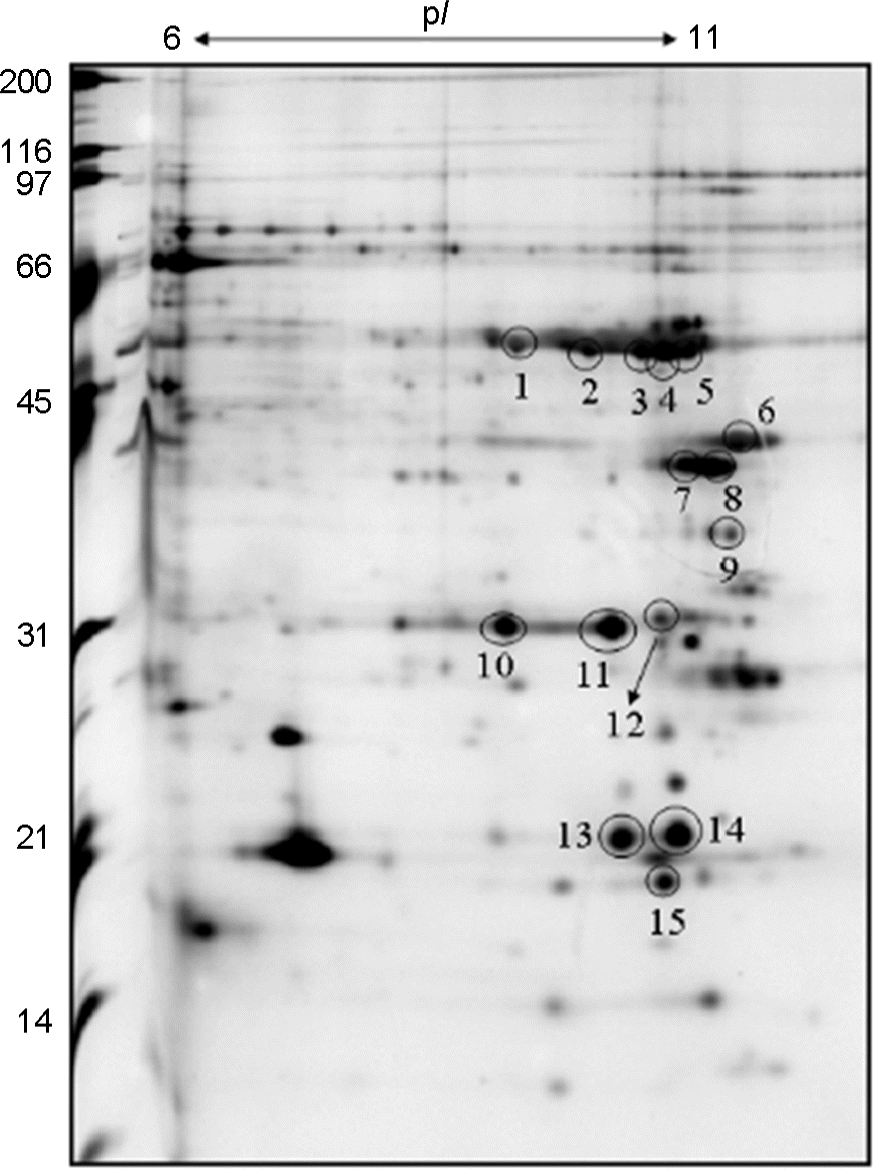 | Figure 1.
H. pylori strain 51 outer membrane protein 2-DE map. The sarcosyl-insoluble proteins were separated on an IPG strip and detected via silver staining. The proteins of the circled spots were identified by Q-TOF. |
Table 1.
Identification of H. pylori 51 OMPs by amino acid sequence analysis




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download