Abstract
Shiga toxin-producing Escherichia coli (STEC) can cause a broad spectrum of human illness ranging from symptom-free to hemolytic uremic syndrom (HUS). Associations between known or putative virulence factors of STEC and diseases in human were investigated. PCR analyses showed that 33 (78.6%) isolates carried an ehxA enterohemolysin gene and 6 (14.3%) isolates possessed an saa autoaggutinating adhesin gene, and 31 (73.8%) isolates carried an eae intimin gene (7 isolates with type β, 16 with type γ, and 3 with type ε). Twenty-nine (69%) isolates from patients carried eae+, ehxA+, saa- (genotype A) and 68 (86%) isolates from asymptomatic outbreaks and 4 (36%) isolates from bovine possessed eae-, ehxA+, saa+ (genotype C). Neither the bundle-forming pilus gene nor the enteropathogenic E. coli adherence factor plasmid was found. In HEp-2 cell adherence assay, isolates carrying eae gene exhibited a localized adherence phenotype, the other isolates carrying saa showed LC (loose clusters of bacteria) and IS (isolated bacteria). In conclusion, most STEC isolated from cattle feces in Gwangju, Korea showed characteristics different from those isolated from patients. But these results may be useful information for pathogenesis judgement of STEC.
Go to : 
REFERENCES
1). Griffin PM., Tauxe RV. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli. and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991. 13:60–98.
2). Levine MM., Xu JG., Kaper JB., Lior H., Prado V., Tall B., Nataro J., Karch H., Wachsmuth K. A DNA probe to identify enterohemorrhagic Escherichia coli of O157:H7 and other serotypes that cause hemorrhagic colitis and hemolytic uremic syndrome. J Infect Dis. 1987. 156:175–82.
3). Gyles CL. Escherichia coli cytotoxins and enterotoxins. Can J Microbiol. 1992. 38:734–46.
4). Finlay BB., Rosenshine I., Donnenberg MS., Kaper JB. Cytoskeletal composition of attaching and effacing lesions associated with enteropathogenic Escherichia coli adherence to Hela cells. Infect Immun. 1992. 60:2541–3.
5). Jerse AE., Yu J., Tall BD., Kaper JB. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA. 1990. 87:7839–43.
6). Paton AW., Srimanote P., Woodrow MC., Paton JC. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect Immun. 2001. 69:6999–7009.
7). Srimanote P., Paton AW., Paton JC. Characterization of a novel type IV pilus locus encoded on the large plasmid of locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect Immun. 2002. 70:3094–100.
8). Boerlin P., McEwen SA., Boerlin-Petzold F., Wilson JB., Johnson RP., Gyles CL. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J Clin Microbiol. 1999. 37:497–503.
9). Pradel N., Bertin Y., Martin C., Livrelli V. Molecular analysis of Shiga toxin-producing Escherichia coli strains isolated from hemolytic uremic syndrome patients and dairy samples in France. Appl Environ Microbiol. 2008. 74:2118–28.
10). Brunder W., Schmidt H., Karch H. KatP. a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 1996. 142:3305–15.
11). Schmidt H., Henkel B., Karch H. A gene cluster closely related to type II secretion pathway operons of gram-negative bacteria is located on the large plasmid of enterohemorrhagic Escherichia coli O157 strains. FEMS Microbiol Lett. 1997. 148:265–72.
12). Brunder W., Schmidt H., Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157: H7 cleaves human coagulation factor V. Mol Microbiol. 1997. 24:767–78.
13). Kim Y-B. Studies on virulence factors and application of arbitrarily-primed polymerase chain reaction analysis to epidemiological of Escherichia coli O157:H7. J Bacteriol Virol. 2001. 31:123–31.
14). Paton AW., Paton JC. Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J Clin Microbiol. 2002. 40:271–4.
15). China B., Goffaux F., Prison V., Mainil J. Comparison of eae, tir, espA and espB genes of bovine and human attaching and effacing Escherichia coli by multiplex polymerase chain reaction. FEMS Microbiol Lett. 1999. 178:177–82.
16). Goffaux F., China B., Janssen L., Mainil J. Genotypic characterization of enteropathogenic Escherichia coli (EPEC) isolated in Belgium from dogs and cats. Res Microbiol. 2001. 151:865–71.
17). McNally A., Roe AJ., Sompson S., Thomson-Carter FM., Hoey DE., Currie C., Chakraborty T., Smith DG., Gally DL. Differences in levels of secreted locus of enterocyte effacement proteins between human disease-associated and bovine Escherichia coli O157. Infect immun. 2001. 69:5107–14.
18). McDaniel TK., Jarvis KG., Donnenberg MS., Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995. 92:1664–8.

19). Paton AW., Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998. 36:598–602.
20). Kobayashi H., Shimada J., Nakazawa M., Morozumi T., Pohjanvirta T., Pelkonen S., Yamamoto K. Prevalence and characteristics of Shiga toxin-producing Escherichia coli from healthy cattle in Japan. Appl Environ Microbiol. 2001. 67:484–9.
21). Wieler LH., Vieler E., Erpenstein C., Schlapp T., Steinruck H., Bauerfeind R., Byomi A., Baljer G. Shiga toxin-producing Escherichia coli strains from bovines: association of adhesion with carriage of eae and other genes. J Clin Microbiol. 1996. 34:2980–4.
22). Franke J., Franke S., Schmidt H., Schwarzkopf A., Wieler LH., Baljer G., Beutin L., Karch H. Nucleotide sequence analysis of enteropathogenic Escherichia coli (EPEC) adherence factor probe and development of PCR for rapid detection of EPEC harboring virulence plasmids. J Clin Microbiol. 1994. 32:2460–3.
23). Cravioto A., Gross RJ., Scotland SM., Rowe B. An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Curr Microbiol. 1979. 3:95–9.
24). Paton AW., Srimanote P., Woodrow MC., Paton JC. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect Immun. 2001. 69:6999–7009.
25). Paton AW., Woodrow MC., Doyle RM., Lanser JA., Paton JC. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J Clin Microbiol. 1999. 37:3357–61.
26). Toma C., Martinez Espinosa E., Song T., Miliwebsky E., Chinen I., Iyoda S., Iwanaga M., Rivas M. Distribution of putative adhesins in different seropathotypes of Shiga toxin-producing Escherichia coli. J Clin Microbiol. 2004. 42:4937–46.
27). Moon HW., Whipp SC., Argenzio RA., Levine MM., Giannella RA. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect Immun. 1983. 41:1340–51.
28). Adu-Bobie J., Frankel G., Bain C., Goncalves AG., Trabulsi LR., Douce G., Knutton S., Dougan G. Detection of intimins alpha, beta, gamma, and delta, four intimin derivatives expressed by attaching and effacing microbial pathogens. J Clin Microbiol. 1998. 36:662–8.
29). Oswald E., Schmidt H., Morabito S., Karch H., Marche's O., Caprioli A. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect Immun. 2000. 68:64–71.
30). Frankel G., Candy DC., Fabiani E., Adu-Bobie J., Gil S., Novakova M., Phillips AD., Dougan G. Molecular characterization of a carboxy-terminal eukaryotic-cell-binding domain of intimin from enteropathogenic Escherichia coli. Infect Immun. 1995. 63:4323–8.
31). Gyles CL. Shiga toxin-producing Escherichia coli: An overview. J Anim Sci. 2007. 85(13 Suppl.):E45-62.
32). Bertin Y., Boukhors K., Livrelli V., Martin C. Localization of the insertion site and pathotype determination of the locus of enterocyte effacement of Shiga toxin-producing Escherichia coli strains. Appl Environ Microbiol. 2004. 70:61–8.
33). Garrido P., Blanco M., Moreno-Paz M., Briones C., Dahbi G., Balnco J., Balnco J., Parro V. STEC-EPEC oligonucleotide microarray: A new tool for typing genetic variants of the LEE pathogenicity island of human and animal Shiga toxin-producing Escherichia coli (STEC) and enteropathogenic E. coli (EPEC) strains. Clin Chem. 2006. 52:192–201.
34). Yuste M., Orden JA., De La Fuente R., Ruiz-Santa-Quiteria JA., Cid D., Martinez-Pulgarin S., Dominquez-Bernal G. Polymerase chain reaction typing of genes of the locus of enterocyte effacement of ruminant attaching and effacing Escehrichia coli. Can J Vet Res. 2008. 72:444–8.
35). Schmidt H., Zhang WL., Hemmrich U., Jelacic S., Brunder W., Tarr PI., Dobrindt U., Hacker J., Karch H. Identification and characterization of a novel genomic island integrated at selC in locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli. Infect Immun. 2001. 69:6863–73.
36). Karch H., Bielaszewska M. Sorbitol-fermenting shiga toxin-producing Escherichia coli O157:H(–) strains: epidemiology, phenotypic and molecular characteristics, and microbiological diagnosis. J Clin Microbiol. 2001. 39:2043–9.
37). Pradel N., Boukhors K., Bertin Y., Forestier C., Martin C., Livrelli V. Heterogeneity of Shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients, cattle, and food samples in central France. Appl Environ Microbiol. 2001. 67:2460–8.
38). Vieira MA., Andrade JR., Trabulsi LR., Rosa AC., Dias AM., Rames SR., Frankel G., Gomes TA. Phenotypic and genotypic characteristics of Escherichia coli strains of non-enteropathogenic E. coli (EPEC) serogroups that carry EAE and lack the EPEC adherence factor and Shiga toxin DNA probe sequence. J Infect Dis. 2001. 183:762–72.
39). Bokete TN., Whittam TS., Wilson RA., Clausen CR., O'Callahan CM., Moseley SL., Fritsche TR., Tarr PI. Genetic and phenotypic analysis of Escherichia coli with enteropathogenic characteristics isolated from Seattle children. J Infect Dis. 1997. 175:1382–9.
40). Scotland SM., Smith HR., Rowe B. Escherichia coli O128 strains from infants with diarrhea commonly show localized adhesion and positivity in the fluorescent-actin staining test but do not hybridize with an enteropathogenic E. coli adherence factor probe. Infect Immun. 1991. 59:1569–71.
41). Gordillo ME., Reeve GR., Pappas J., Mathewson JJ., DuPunt HL., Murray BE. Molecular characterization of strains of enteroinvasive Escherichia coli O143, including isolates from a large outbreak in Houston, Texas. J Clin Microbiol. 1992. 30:889–93.
42). Roe AJ., Hoey DE., Gally DL. Regulation, secretion and activity of type III-secreted proteins of enterohaemorrhagic Escherichia coli O157. Biochem Soc Trans. 2003. 31:98–103.
43). Tobe T., Sasakawa C. Role of bundle-forming pilus of enteropathogenic Escherichia coli in host cell adherence and in microcolony development. Cell Microbiol. 2001. 3:579–85.
44). Knutton S., Shaw RK., Anantha RP., Donnenberg MS., Zorgani AA. The type IV bundle-forming pilus of enteropathogenic Escherichia coli undergoes dramatic alterations in structure associated with bacterial adherence, aggregation and dispersal. Mol Microbiol. 1999. 33:499–509.
45). Tatsuno I., Horie M., Abe H., Miki T., Makino K., Shinagawa H., Taguchi H., Kamiya S., Hayashi T., Sasakawa C. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect Immun. 2001. 69:6660–9.
46). Brunder W., Khan AS., Hacker J., Karch H. Novel type of fimbriae encoded by the large plasmid of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H(–). Infect Immun. 2001. 69:4447–57.
47). Tarr PI., Bilge SS., Vary JC Jr., Jelacic S., Habeeb RL., Ward TR., Baylor MR., Besser TE. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect Immun. 2000. 68:1400–7.
48). Nicholls L., Grant TH., Robins-Browne RM. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol Microbiol. 2000. 35:275–88.
49). Doughty S., Sloan J., Bennett-Wood V., Robertson M., Robins-Browne RM., Hartland EL. Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect Immun. 2002. 70:6761–9.
50). Torres AG., Giron JA., Perna NT., Burland V., Blattner FR., Avelino-Flores F., Kaper JB. Identification and characterization of lpfABCC'DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2002. 70:5416–27.
51). Zhang WL., Kohler B., Oswald E., Beutin L., Karch H., Morabito S., Caprioli A., Suerbaum S., Schmidt H. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J Clin Microbiol. 2002. 40:4486–92.
Go to : 
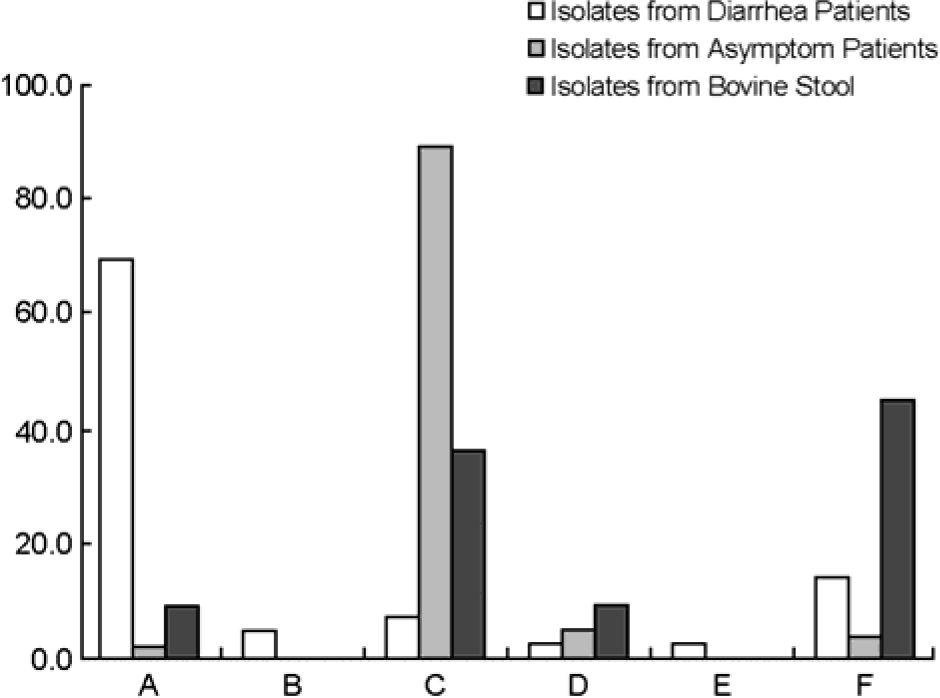 | Figure 2.Distribution of the STEC genotype by multiplex PCR. A: eaeA+, ehxA+, saa-, B: eaeA+, ehxA-, saa-, C: eaeA-, ehxA+, saa+, D: eaeA-, ehxA+, saa-, E: eaeA-, ehxA-, saa+, F: eaeA-, ehxA-, saa- (+: gene present; -: gene absent). |
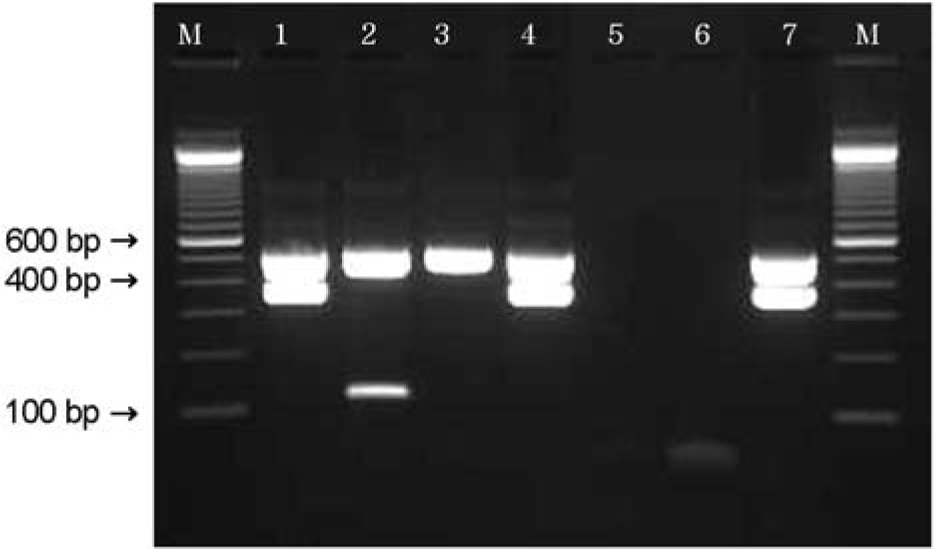 | Figure 3.Multiplex PCR analysis of the saa (119 bp), eae (384 bp), the ehxA (534 bp) genes. M: 100 bp ladder, lane 1: genotype A (isolate GJ-04-08-23), lane 2: genotype C (isolate Jinwol-SBM), lane 3: genotype D (isolate bovine 20), lane 4: genotype A (isolate GJ-08-07-200), lane 5: genotyep F (isolate bovine 31), lane 6: negative control, lane 7: positive control (EDL 933). |
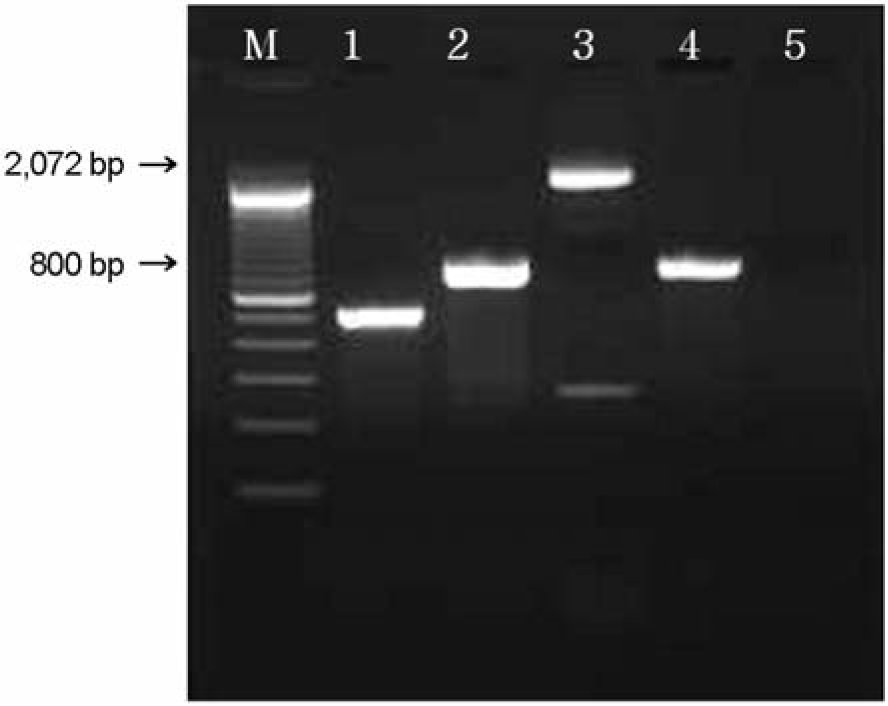 | Figure 4.PCR result for the detection of eaeβ (520 bp), eaeγ (778 bp) and eaeε (2,069 bp) genes. M: 100 bp ladder, lane 1: eaeβ + (isolate GJ-04-07-84), lane 2: eaeγ + (isolate GJ-05-06-290), lane 3: eaeε + (isolate GJ-05-09-32), lane 4: positive control (EDL 933), lane 5: negative control. |
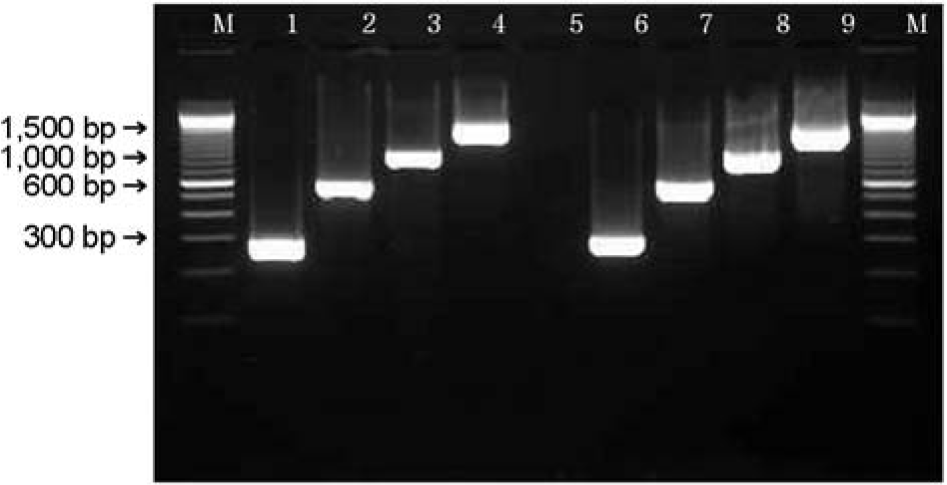 | Figure 5.PCR result for the detection of espA (299 bp), espB (633 bp), espD (939 bp) and tir (1,550 bp) genes. M: 100 bp ladder, lane 1~4: espA +, espB +, espD +, tir + (isolate GJ-05-09-032), lane 5: negative control, lane 6~9: positive control (EDL 933). |
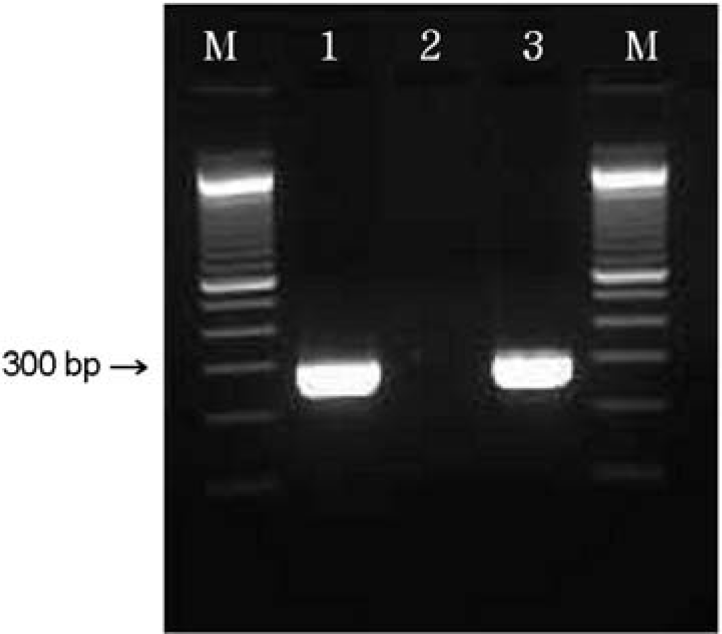 | Figure 6.PCR analysis of the espP (301 bp)genes. M: 100 bp ladder, lane 1: espP + (isolate GJ-07-06-324), lane 2: negative control, lane 3: positive control (EDL 933). |
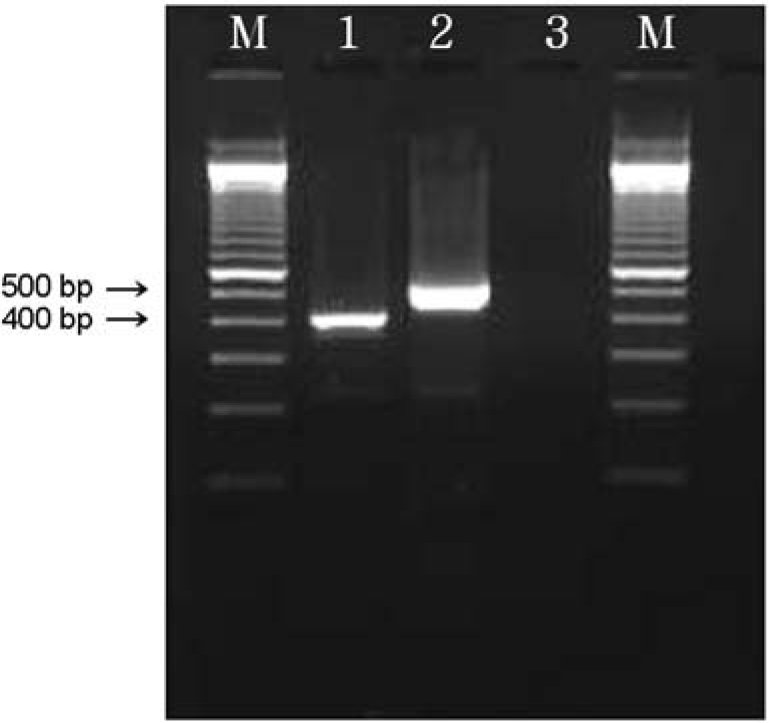 | Figure 7.Multiplex PCR to distinguish an intact selC locus from one disrupted by LEE. M: 100 bp ladder, lane 1: disrupted selC (418 bp, isolate GJ-05-06-150), lane 2: Intact selC (527 bp, isolate GJ-05-06-290), lane 3: ND (not detected). |
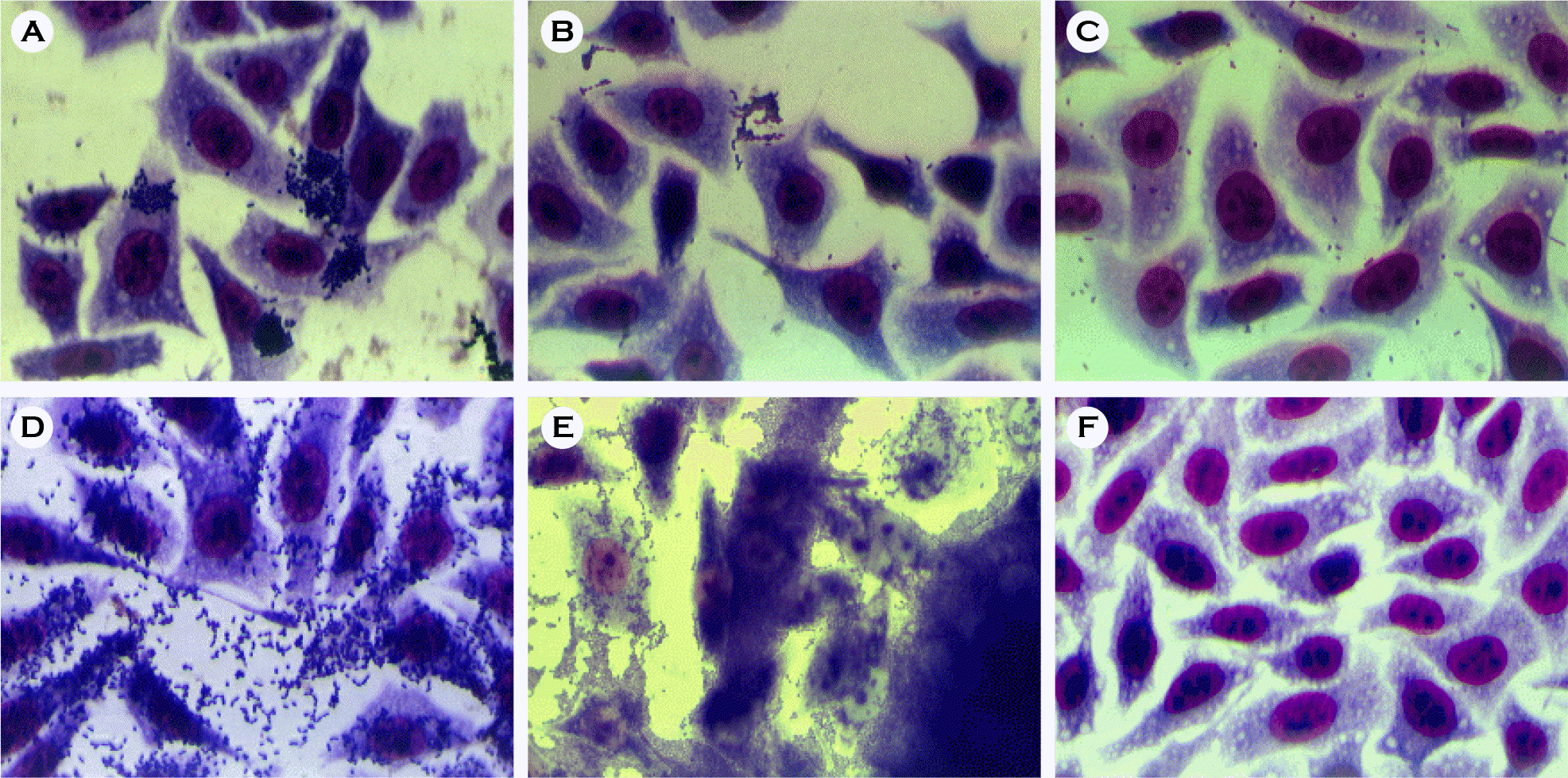 | Figure 8.Adherence patterns of STEC in HEp-2 cell culture. A: LA pattern, isolate GJ-04-07-84, B: LC pattern, isolate Jinwol 9, C: IS pattern: isolate Jinwol SBM, D: DA pattern, isolate bovine 27, E: AA pattern, isolate GJ-08-07-54, F: Negative control. |
Table 1.
PCR primers to amplify specific fragments from the various pathogenic genes in STEC
Table 2.
Distribution of the virulence factor-encoding genes according to symptoms of the STEC isolatesa
| Geneb | No. (%) of isolates | P valuec | |
|---|---|---|---|
| Asymptomatic | Diarrhea patients | ||
| saa | 70 (88.6) | 6 (14.3) | <0.0001 |
| eae | 4 (5.1) | 31 (73.8) | <0.0001 |
| ehxA | 74 (93.7) | 33 (78.6) | NS (0.7803) |
| espP | 6 (50) | 26 (62) | NS (0.5173) |
Table 3.
Virulence factors of the 67 STEC strains tested
| Strain | Origin | Age | Sex | Disease Associationa | Sero Groupb | Sorbitol | stx1 | stx2 | RPLA | saa | ehxA | Eaec | tir | espA | espB | espD | espP | bfpA | EAF | SelCd | Adherence pattern |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 27-1 | Bovine | – | – | – | NT | – | + | – | + | + | + | – | – | – | – | – | – | – | – | I | AA |
| 128 | Bovine | – | – | – | NT | – | – | + | – | – | – | – | – | – | – | – | – | – | – | ND | IS |
| 153 | Bovine | – | – | – | O113 | – | – | + | + | + | + | – | – | – | – | – | – | – | – | I | LC |
| 126 | Bovine | – | – | – | O117 | – | + | + | + | – | – | – | – | – | – | – | + | – | – | ND | IS |
| 20 | Bovine | – | – | – | O168 | – | – | + | – | – | + | – | – | – | – | – | – | – | – | I | NA |
| 27 | Bovine | – | – | – | O178 | – | + | + | + | + | + | – | – | – | – | – | – | – | – | D | DA |
| 174 | Bovine | – | – | – | O2 | – | – | + | – | – | – | – | – | – | – | – | + | – | – | D | IS |
| 31 | Bovine | – | – | – | O22 | – | – | + | – | – | – | – | – | – | – | – | – | – | – | D | NA |
| 171 | Bovine | – | – | – | O46 | – | – | + | – | – | – | – | – | – | – | – | – | – | – | I | IS |
| 180 | Bovine | – | – | – | O5 | – | – | + | – | – | + | +(UT) | – | – | – | – | – | – | – | D | IS |
| 189 | Bovine | – | – | – | O6 | – | – | + | + | + | + | – | – | – | – | – | + | – | – | D | NA |
| GJ-06-07-117 | Human | 1 | M | D | NT | – | + | – | + | – | + | +(β) | – | – | + | + | + | – | – | ND | LA |
| GJ-05-06-150 | Human | 7 | M | D | O103 | – | + | – | + | – | + | +(UT) | – | – | – | – | – | – | – | D | LA |
| GJ-05-08-057 | Human | 2 | M | D | O103 | – | + | – | + | – | + | +(UT) | – | – | – | – | – | – | – | ND | LA |
| GJ-06-07-222 | Human | 2 | F | D | O103 | – | + | – | + | – | + | +(UT) | + | – | + | + | – | – | – | I | LA |
| GJ-05-06-290 | Human | 9 | F | D | O111 | – | + | + | + | – | – | +(γ) | + | – | + | + | + | – | – | I | LC |
| GJ-05-11-087 | Human | 1 | M | D | O111 | – | + | – | + | – | + | +(γ) | – | – | + | + | + | – | – | I | LA |
| GJ-07-07-066 | Human | 0 | M | D | O111 | – | + | + | + | – | + | +(γ) | – | – | – | – | + | – | – | I | LA |
| GJ-07-08-056 | Human | 1 | M | D | O111 | – | + | – | + | – | + | +(γ) | – | – | + | – | + | – | – | I | NA |
| GJ-08-07-200 | Human | 5 | M | D | O111 | – | + | + | + | – | + | +(γ) | – | – | + | + | + | – | – | I | IS |
| GJ-08-07-422 | Human | 1 | M | D | O111 | – | + | – | + | – | + | +(γ) | – | – | – | – | + | – | – | I | LA |
| GJ-08-08-147 | Human | 1 | M | D | O111 | – | + | – | + | – | + | +(γ) | – | – | – | – | + | – | – | I | LC |
| GJ-07-08-040 | Human | 0 | F | D | O112ac | – | – | + | + | – | – | – | – | – | – | – | – | – | – | ND | IS |
| GJ-08-10-091 | Human | 1 | F | D | O117 | – | + | – | + | – | + | +(UT) | + | – | + | + | – | – | – | I | LA |
| GJ-08-07-100 | Human | 8 | M | D | O120 | – | – | + | + | – | – | – | – | – | – | – | – | – | – | I | IS |
| GJ-05-07-165 | Human | 2 | M | D | O127 | – | + | – | + | – | + | +(β) | + | – | + | + | – | – | – | ND | AA |
| GJ-08-07-205 | Human | 5 | M | D | O128 | – | + | – | + | – | – | – | – | – | – | – | – | – | – | I | NA |
| GJ-04-08-23 | Human | 2 | F | D | O145 | – | + | – | + | – | + | +(γ) | + | + | + | + | + | – | – | I | LA |
| GJ-05-09-032 | Human | 1 | F | D | O145 | – | + | – | + | – | + | +(ε) | + | + | + | + | + | – | – | I | IS |
| GJ-05-11-177 | Human | 1 | F | D | O157 | + | – | + | + | – | + | +(γ) | – | + | + | + | + | – | – | ND | IS |
| GJ-07-06-324 | Human | 1 | M | D | O157 | + | + | + | + | – | + | +(γ) | – | + | + | + | + | – | – | D | IS |
| GJ-07-07-088 | Human | 2 | M | D | O157 | + | + | + | + | – | + | +(γ) | – | + | + | + | + | – | – | ND | IS |
| GJ-07-07-216 | Human | 1 | F | D | O157 | + | + | + | + | – | + | +(γ) | + | + | + | + | + | – | – | I | LA |
| GJ-07-08-210 | Human | 1 | F | D | O157 | + | + | + | + | – | + | +(γ) | + | + | + | + | + | – | – | ND | NA |
| GJ-08-05-148 | Human | 67 | M | D | O157 | + | + | + | + | – | + | +(γ) | + | + | + | + | + | – | – | D | IS |
| GJ-05-09-110 | Human | 2 | M | D | O166 | – | + | – | + | – | + | +(γ) | + | – | + | + | – | – | – | ND | IS |
| GJ-05-07-078 | Human | 8 | M | D | O168 | – | – | + | – | – | + | +(β) | – | – | – | – | – | – | – | D | IS |
| GJ-04-07-84 | Human | 1 | F | D | O26 | – | + | + | + | – | + | +(β) | + | – | + | + | + | – | – | I | LA |
| GJ-05-07-037 | Human | 2 | M | D | O26 | – | + | + | + | – | – | +(β) | + | – | + | + | + | – | – | ND | LC |
| GJ-05-07-164 | Human | 4 | F | D | O26 | – | + | – | + | – | + | +(UT) | – | – | – | – | + | – | – | I | LA |
| GJ-05-10-167 | Human | 3 | F | D | O26 | – | + | – | + | – | + | +(β) | + | – | + | + | + | – | – | ND | LA |
| GJ-07-05-219 | Human | 44 | F | D | O26 | – | + | – | + | – | + | +(β) | + | – | + | + | + | – | – | ND | LA |
| GJ-08-07-438 | Human | 6 | M | D | O55 | – | + | – | – | + | + | – | – | – | – | – | + | – | – | I | IS |
| GJ-07-09-032 | Human | 11 | M | D | O84 | – | + | – | – | – | – | – | – | – | – | – | – | – | – | ND | LC |
| Gwangsan 3 | Human | – | F | SF | NT | – | – | + | + | + | + | – | – | – | – | – | + | – | – | ND | LC |
| Jinwol 1071 | Human | – | – | SF | NT | – | + | – | – | – | + | – | – | – | – | – | – | – | – | ND | IS |
| Yongdoo 29 | Human | – | – | SF | O103 | – | + | – | + | – | + | +(ε) | + | – | + | + | – | – | – | I | LA |
| KBM | Human | – | M | SF | O103 | – | + | – | + | – | + | +(ε) | + | – | + | + | – | – | – | I | LC |
| Jinwol 1682 | Human | – | – | SF | O115 | – | + | – | + | – | + | – | – | – | – | – | – | – | – | I | LC |
| Yongdoo 26 | Human | – | – | SF | O141 | – | – | + | + | – | – | – | – | – | – | – | – | – | – | ND | NA |
| Jinwol 457 | Human | – | – | SF | O146 | – | – | + | + | + | + | – | – | – | – | – | + | – | – | I | IS |
| Jinwol 1692 | Human | – | – | SF | O146 | – | – | + | – | – | + | – | – | – | – | – | – | – | – | D | NA |
| KIW | Human | – | F | SF | O157 | + | + | + | + | – | + | +(γ) | + | + | + | + | + | – | – | D | IS |
| Jinwol 242 | Human | – | – | SF | O163 | – | – | + | + | + | + | – | – | – | – | – | – | – | – | ND | NA |
| DG3 | Human | – | F | SF | O168 | – | – | + | + | – | + | – | – | – | – | – | – | – | – | I | NA |
| Gwangsan5 | Human | – | F | SF | O174 | – | – | + | + | + | + | – | – | – | – | – | + | – | – | I | IS |
| Jinwol 41 | Human | – | – | SF | O26 | – | + | – | + | – | + | +(UT) | + | – | + | + | + | – | – | ND | LA |
| Yongdoo 30 | Human | – | – | SF | O84 | – | + | – | – | – | – | – | – | – | – | – | – | – | – | ND | NA |
| Gwangsan3-2 | Human | – | F | SF | O91 | – | + | – | + | + | + | – | – | – | – | – | + | – | – | D | LC |
| Jinwol-SBM | Human | – | F | SF | O91 | – | + | + | + | + | + | – | – | – | – | – | + | – | – | D | IS |
| Jinwol 9 | Human | – | – | SF | O91 | – | + | + | + | + | + | – | – | – | – | – | + | – | – | D | IS |
| Jinwol 13 | Human | – | – | SF | O91 | – | + | + | + | + | + | – | – | – | – | – | + | – | – | D | LC |
| Jinwol 1222 | Human | – | – | SF | O91 | – | + | + | + | – | – | – | – | – | – | – | – | – | – | D | IS |
| Jinwol 1269 | Human | – | – | SF | O91 | – | + | + | + | – | – | – | – | – | – | – | – | – | – | D | IS |
| Jinwol 1672 | Human | – | – | SF | O91 | – | + | – | + | – | + | – | – | – | – | – | + | – | – | ND | AA |
| pork | food | – | – | – | NT | – | – | + | – | – | + | – | – | – | – | – | + | – | – | I | IS |
| beef | food | – | – | – | O116 | – | + | + | + | + | + | – | – | – | – | – | + | – | – | ND | IS |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download