Abstract
Japanese encephalitis virus (JEV), a member of mosquito-borne flaviviruses, is the leading cause of viral encephalitis in a large geographic area of Southeast Asia and Australia. JEV contains a single-stranded positive-sense RNA genome, which encodes its own RNA-dependent RNA polymerase (NS5) that is required for genomic RNA replication. In this study, we have described a pair of mouse antisera specific to the N- or C-terminal region of the NS5. Initially, two hydrophilic regions corresponding to the N-terminus and C-terminus of the NS5 protein were individually amplified by reverse transcription-PCR from the genomic RNA of JEV K87P39 strain. The amplified DNA fragments were cloned into a prokaryotic expression vector, pGEX-4T-1; the resulting constructs were used for the expression of GST fusion proteins, designated GST/NS5N and GST/NS5C, in E. coli BL-21 strain. Following immunization of three BALB/c mice with each of the purified GST/NS5N and GST/NS5C, we obtained two pools of the antisera, specifically recognizing the ~103-kDa NS5 and several smaller NS5-related proteins in BHK-21 and Vero cells infected with JEV K87P39 strain. Overall, we have successfully expressed the N- and C-terminal regions of JEV NS5 fused to the C-terminus of GST and generated the mouse antisera capable of recognizing the NS5 and its related proteins in JEV-infected cells. This would provide a valuable reagent for the study of JEV NS5 in the viral life cycle.
Go to : 
REFERENCES
1). Burke DS., Leake CJ. 1988. Japanese encephalitis. In. Monath TP, editor. (ed.),. The Arboviruses: Epidemiology and Ecology. III:pp.p. 63–92. CRC Press;Florida:

2). Vaughn DW., Hoke CH Jr. The epidemiology of Japanese encephalitis: prospects for prevention. Epidemiol Rev. 1992. 14:197–221.

3). Tsai TF. New initiatives for the control of Japanese encephalitis by vaccination: Minutes of a WHO/CVI meeting, Bangkok, Thailand, 13-15 October 1998. Vaccine. 2000. 18:1–25.
4). Hashimoto H., Nomoto A., Watanabe K., Mori T., Takezawa T., Arizawa C., Takegami T., Hiramatsu K. Molecular cloning and complete nucleotide sequence of the genome of Japanese encephalitis virus Beijing-1 strain. Virus Genes. 1988. 1:305–17.

5). World Health Organization Japanese encephalitis vaccines. Wkly Epidemiol Rec. 1998. 73:334–44.
6). Hanna JN., Ritchie SA., Phillips DA., Lee JM., Hills SL., van den Hurk AF., Pyke AT., Johansen CA., Mackenzie JS. Japanese encephalitis in north Queensland, Australia. 1998. Med J Aust. 1999. 170:533–6.

7). Mackenzie JS., Poidinger M., Phillips DA., Johansen CA., Hall RA., Hanna JN., Ritchie SA., Shield J., Graham R. 1997. Emergence of Japanese encephalitis virus in the Australian region. In. Saluzzo J, Dodet B, editors. (eds.),. Factors in the Emergence of Arboviruses Diseases. pp.p. 191–201. Elsevier;Paris:
8). Shield J., Hanna JN., Phillips DA. Reappearance of the Japanese encephalitis virus in the Torres Strait. Comm Dis Intel. 1996. 20:191.
9). Spicer PE., Phillips D., Pike A., Johansen C., Melrose W., Hall RA. Antibodies to Japanese encephalitis virus in human sera collected from Irian Jaya: Follow-up of a previously reported case of Japanese encephalitis in that region. Trans R Soc Trop Med Hyg. 1999. 93:511–4.

10). Rice CM. 1996. Flaviviridae: The viruses and their replication. In. Fields BN, Knipe DM, Howley PM, editors. (eds.),. Fields Virology. 3rd ed. pp.p. 931–960. Lippincott-Raven Publishers;Philadelphia:
11). Thiel HJ., Plagemann PGW., Moennig V. 1996. Pestiviruses. In. Fields BN, Knipe DM, Howley PM, editors. (eds.),. Fields Virology. 3rd ed. pp.p. 1059–1073. Lippincott-Raven Publishers;Philadelphia:
12). Burke DS., Monath TP. 2001. Flaviviruses. In. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. (eds.),. Fields Virology. 4th ed. pp.p. 1043–1125. Lippincortt Williams & Wilkins;Philadelphia:
13). Lindenbach BD., Rice CM. 2001. Flaviviridae: The viruses and their replication. In. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. (eds.),. Fields Virology. 4th ed. pp.p. 991–1041. Lippincortt Williams & Wilkins;Philadelphia:
14). Chambers TJ., Hahn CS., Galler R., Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990. 44:649–88.

15). Egloff MP., Benarroch D., Selisko B., Romette JL., Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002. 21:2757–68.

16). Koonin EV. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J Gen Virol. 1993. 74:733–40.
17). Ackermann M., Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J Biol Chem. 2001. 276:39926–37.

18). Guyatt KJ., Westaway EG., Khromykh AA. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J Virol Methods. 2001. 92:37–44.

19). Tan BH., Fu J., Sugrue RJ., Yap EH., Chan YC., Tan YH. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology. 1996. 216:317–25.
20). Medin CL., Fitzgerald KA., Rothman AL. Dengue virus nonstructural protein NS5 induces interleukin-8 transcription and secretion. J Virol. 2005. 79:11053–61.

21). Best SM., Morris KL., Shannon JG., Robertson SJ., Mitzel DN., Park GS., Boer E., Wolfinbarger JB., Bloom ME. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J Virol. 2005. 79:12828–39.

22). Yun SI., Kim SY., Choi WY., Nam JH., Ju YR., Park KY., Cho HW., Lee YM. Molecular characterization of the full-length genome of the Japanese encephalitis viral strain K87P39. Virus Res. 2003. 96:129–40.

23). Sambrook J., Fritsch EF., Maniatis T. 1989. Molecular cloning: a laboratory manual. second ed.Cold Spring Harbor Laboratory;New York:
24). Ray D., Shah A., Tilgner M., Guo Y., Zhao Y., Dong H., Deas TS., Zhou Y., Li H., Shi PY. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol. 2006. 80:8362–70.

25). Zhou Y., Ray D., Zhao Y., Dong H., Ren S., Li Z., Guo Y., Bernard KA., Shi PY., Li H. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007. 81:3891–903.

26). Dong H., Ray D., Ren S., Zhang B., Puig-Basagoiti F., Takagi Y., Ho CK., Li H., Shi PY. Distinct RNA elements confer specificity to flavivirus RNA cap methylation events. J Virol. 2007. 81:4412–21.

27). Alvarez DE., De Lella Ezcurra AL., Fucito S., Gamarnik AV. Role of RNA structures present at the 3'UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005. 339:200–12.

Go to : 
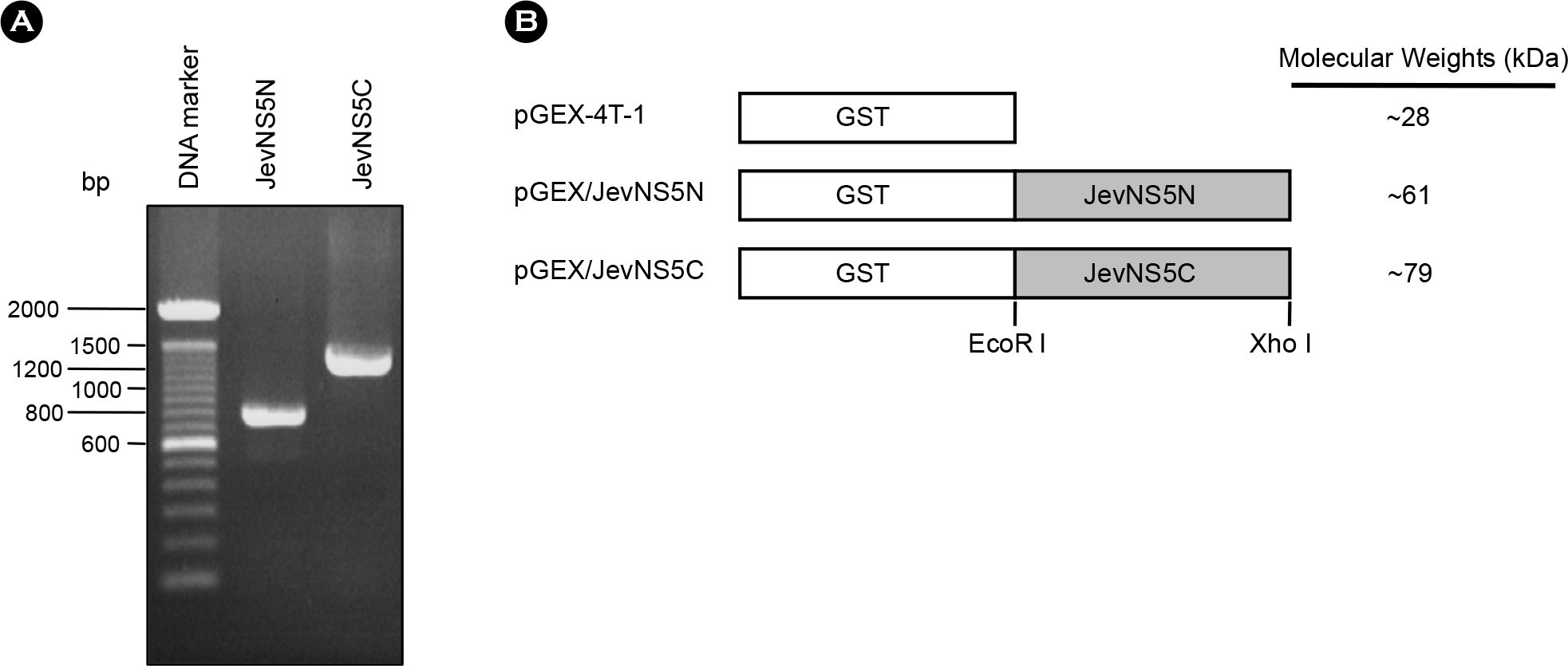 | Figure 1.PCR amplicons of JevNS5N and JevNS5C (A) and schematic presentation of pGEX/JevNS5N and pGEX/JevNS5C constructs (B). The JEV NS5N and NS5C regions were PCR-amplified with an appropriate pair of primers as indicated in the text. The amplicons were separated by 1% agarose gel electrophoresis and stained with ethidium bromide (A). pGEX/JevNS5N and pGEX/JevNS5C constructs were designed to express the N-terminal region (275 amino acids) and C-terminal region (461 amino acids) of JEV NS5 protein, respectively (B). |
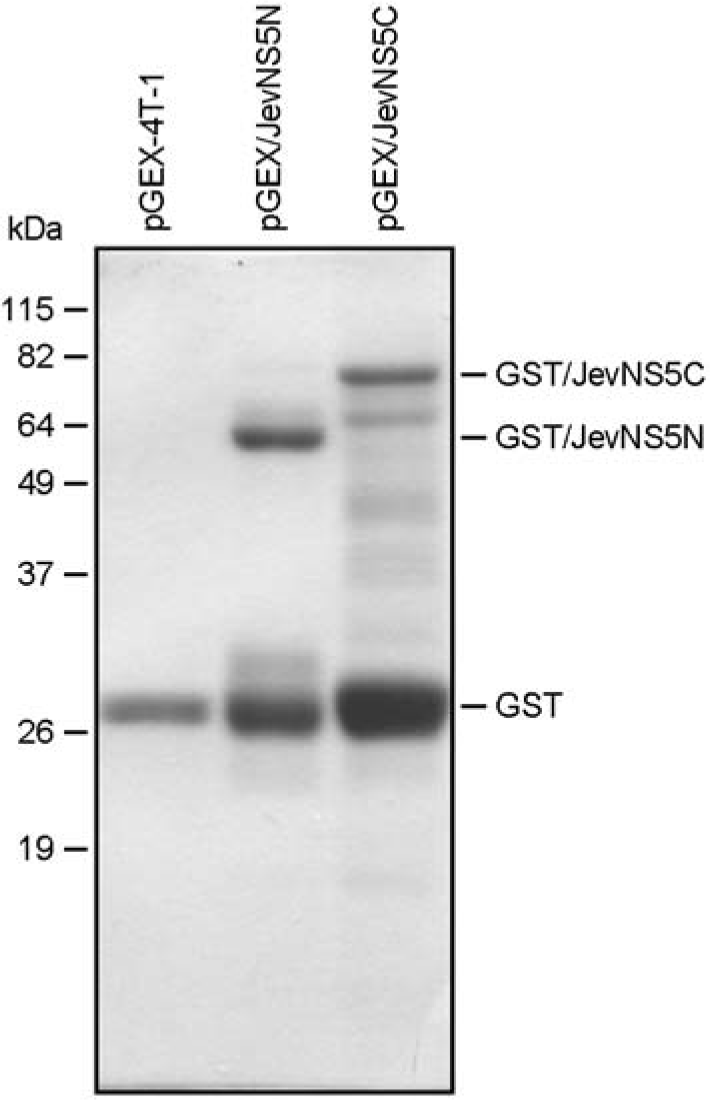 | Figure 2.Expression of GST/JevNS5N and GST/JevNS5C proteins (coommasie staining). The E. coli. BL-21 cells were transformed with the parental pGEX-4T-1 or one of two recombinant plasmids designated pGEX/JevNS5N and pGEX/JevNS5C. The expression of the recombinant proteins (GST/JevNS5N and GST/JevNS5C) was analyzed by 12% SDS-PAGE and coommasie staining. The molecular weight markers in kDa were indicated on the left and the sizes of GST, GST/JevNS5N, and GST/JevNS5C were shown on the right. |
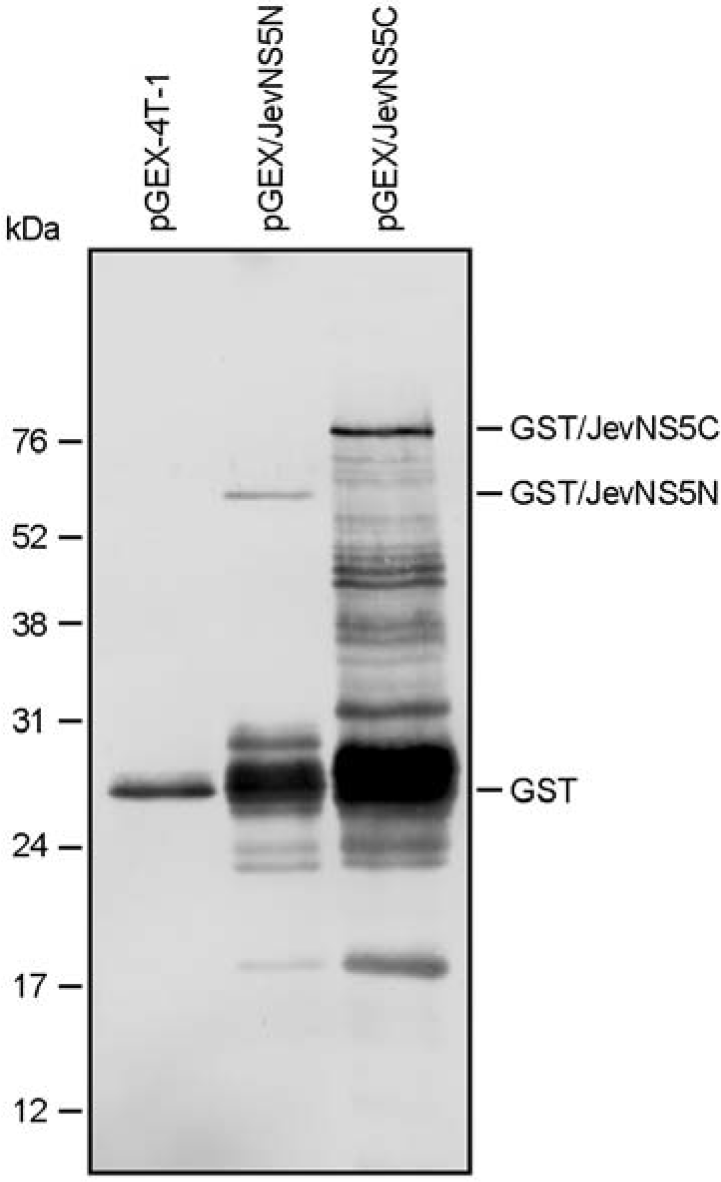 | Figure 3.Identification of GST/JevNS5N and GST/JevNS5C proteins (immunoblotting). The parental pGEX-4T-1 or one of two recombinants (pGEX/JevNS5N and pGEX/JevNS5C) was transformed into E. coli BL-21 cells. Each culture was incubated in the presence of IPTG to induce the GST fusion proteins designated GST/JevNS5N and GST/JevNS5C. Following purification of the recombinant proteins with a glutathione sepharose column, the GST fusion proteins were separated by 12% SDS-PAGE and immuno-staining with a GST-specific mouse monoclonal antibody. |
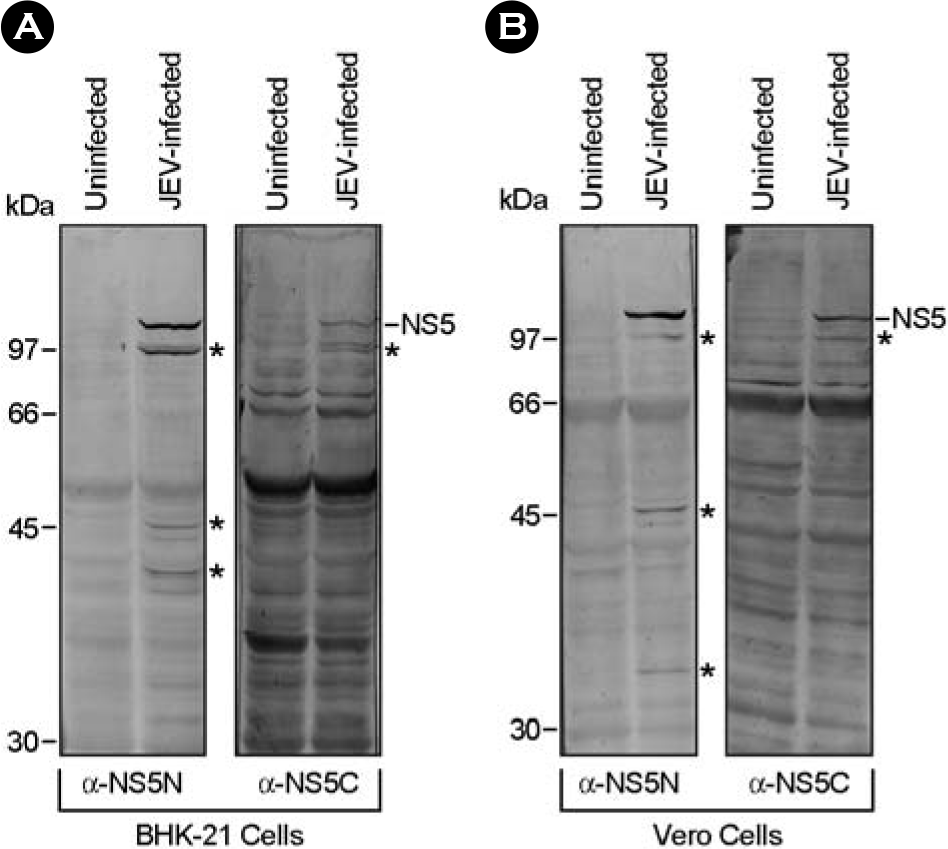 | Figure 4.Immunoblotting with two mouse antisera raised against GST/JevNS5N and GST/JevNS5C. BHK-21 or Vero cells were mock-infected or infected with JEV K87P39 strain at an MOI of 1. At 20 h post-infection, cells were directly lysed with 1 × sample loading buffer and the cell lysates were separated by 12% SDS-PAGE. The NS5 and NS5-related proteins (asterisks) were recognized by two mouse antisera (α-NS5N and α-NS5C) raised against GST/JevNS5N and GST/JevNS5C, respectively. Molecular weight markers (in kDa) were shown on the left and the sizes of NS5 and its related proteins were presented on the right. |
Table 1.
Primers used for PCR amplification in this study




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download