Abstract
To expand the epidemiological understanding of Newcastle disease in Jeju Province, Korea, active surveillance was extensively performed through a virological examination for poultry farms and wild birds in Jeju Province during 2007~2008. Samples (swabs or fresh feces) were collected from a total of 6,485 birds including 6,405 domestic birds (chickens, ducks, pheasants, geese, quails, turkeys, and ostriches) and 80 wild birds. A total of 24 hemagglutinating agents were isolated from domestic birds on fourteen farms including five Korean native chicken, one layer chicken, two broiler chicken, four duck and two pheasant farms. The hemagglutinating agents were all identified as lentogenic NDV based on the reverse transcriptase polymerase chain reaction, sequence analysis of amino acids on the F cleavage site and mean death time in chicken embryos. The F gene-based phylogenetic analysis revealed that the NDV isolates were classified into genotypes 1 or 2 of class II. These lentogenic viruses were closely related to NDV vaccine strains used in Jeju Province. Active surveillance conducted for Newcastle disease indicates no scientific evidence of virulent NDV infection in chickens in Jeju Province, Korea since 2005.
Go to : 
REFERENCES
1). Alexander DJ. Newcatle disease, other avian paramyxoviruses, and pneumovirus infections. Saif YM, Barnes HJ, Glisson JR, Fadly AM, McDougald LR, Swayne DE, editors. editors.Disease of Poultry. 11th ed.Iowa: Iowa State University Press;2003. p. 63–100.
2). Lamb RA., Kolakofsky D. Paramyxoviridae. Knipe DM, Howley PM, editors. editors.Fields Virology. 4th ed.Philadelphia: Lippincott-Raven Publishers;2001. p. 1305–1340.
3). Dutch RE., Hagglund RN., Nagel MA., Paterson RG., Lamb RA. Paramyxovirus fusion (F) protein: a conformational change on cleavage activation. Virology. 2001. 281:138–50.

5). Morrison TG. Structure and function of a paramyxovirus fusion protein. Biochim Biophys Acta. 2003. 1614:73–84.

6). Kim LM., King DJ., Curry PE., Suarez DL., Swayne DE., Stallknecht DE., Slemons RD., Pedersen JC., Senne DA., Winker K., Afonso CL. Phylogenetic diversity among low-virulence Newcastle disease viruses from waterfowl and shorebirds and comparison of genotype distributions to those of poultryorigin isolates. J Virol. 2007. 81:12641–53.

7). Seal BS., Wise MG., Pedersen JC., Senne DA., Alvarez R., Scott MS., King DJ., Yu Q., Kapczynski DR. Genomic sequences of low-virulence avian paramyxovirus-1 (Newcastle disease virus) isolates obtained from live-bird markets in North America not related to commonly utilized commercial vaccine strains. Vet Microbiol. 2005. 106:7–16.

8). Liu XF., Wan HQ., Ni XX., Wu YT., Liu WB. Pathotypical and genotypical characterization of strains of Newcastle disease virus isolated from outbreaks in chicken and goose flocks in some regions of China during 1985-2001. Arch Virol. 2003. 148:1387–403.

9). Liu H., Wang Z., Wu Y., Zheng D., Sun C., Bi D., Zuo Y., Xu T. Molecular epidemiological analysis of Newcastle disease virus isolated in China in 2005. J Virol Methods. 2007. 140:206–11.

10). Aldous EW., Mynn JK., Banks J., Alexander DJ. A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol. 2003. 32:239–56.

11). Lomniczi B., Wehmann E., Herczeg J., Ballagi-Pordany A., Kaleta EF., Werner O., Meulemans G., Jorgensen PH., Mante AP., Gielkens AL., Capua I., Damoser J. Newcastle disease outbreaks in recent years in Western Europe were caused by an old (VI) and a novel genotype (VII). Arch Virol. 1998. 143:49–64.

12). Toyoda T., Sakaguchi T., Imai K., Inocencio NM., Gotoh B., Hamaguchi M., Nagai Y. Structural comparison of the cleavage-activation site of the fusion glycoprotein between virulent and avirulent strains of Newcastle disease virus. Virology. 1987. 158:242–7.

13). World Organisation for Animal Health. Chapter 2.3.14 Newcastle disease. Office International des Epizooties. Manual of diagnostic tests and vaccines for terrestrial animals. 6th ed.Paris: Office International des Epizooties;2008. p. p. 576–589.
14). de Leeuw OS., Koch G., Hartog L., Ravenshorst N., Peeters BP. Virulence of Newcastle disease virus is determined by the cleavage site of the fusion protein and by both the stem region and globular head of the haemagglutinin-neuraminidase protein. J Gen Virol. 2005. 86:1759–69.

15). Garten W., Berk W., Nagai Y., Rott R., Klenk HD. Mutational changes of the protease susceptibility of glycoprotein F of Newcastle disease virus: effects on pathogenicity. J Gen Virol. 1980. 50:135–47.

17). Peeters BP., de Leeuw OS., Koch G., Gielkens AL. Rescue of Newcastle disease virus from cloned cDNA: evidence that cleavability of the fusion protein is a major determinant for virulence. J Virol. 1999. 73:5001–9.

18). Kim JH., Song CS. Consideration of cause of recent severe outbreak of Newcastle disease in Korea and a brief review of virological differences, serological diagnosis and administration of a vaccine. Kor J Poult Sci. 1992. 19:65–76.
19). Kwon HJ., Cho SH., Ahn YJ., Seo SH., Choi KS., Kim SJ. Molecular epidemiology of Newcastle disease in Republic of Korea. Vet Microbiol. 2003. 95:39–48.

20). Lee YJ., Sung HW., Choi JG., Kim JH., Song CS. Molecular epidemiology of Newcastle disease viruses isolated in South Korea using sequencing of the fusion protein cleavage site region and phylogenetic relationships. Avian Pathol. 2004. 33:482–91.

21). World Organisation for Animal Health. Chapter 10.13 Newcastle disease. Office International des Epizooties. Terrestrial animal helath code. 6th ed.Paris: Office International des Epizooties;2008. http://www.oie.int/eng/normes/MCode/en_chapitre_1.10.13.htm.
22). Alexander D. .J.,. 1988b. Newcastle disease diagnosis. Alexander D.J, editor. (Ed.),. Newcastle Disease. Kluwer Academic Publishers;Boston: p. 147–60.

23). Alexander DJ. The epidemiology and control of avian influenza and Newcastle disease. J Comp Pathol. 1995. 112:105–26.

24). Collins MS., Bashiruddin JB., Alexander DJ. Deduced amino acid sequences at the fusion protein cleavage site of Newcastle disease viruses showing variation in antigenicity and pathogenicity. Arch Virol. 1993. 128:363–70.

25). Ke GM., Liu HJ., Lin MY., Chen JH., Tsai SS., Chang PC. Molecular characterization of Newcastle disease viruses isolated from recent outbreaks in Taiwan. J Virol Methods. 2001. 97:1–11.

26). Alexander DJ., Spackman D., Allan WH., Borland L. Isolation of Newcastle disease virus from a wild mallard duck (Anas platyrhynchos). Vet Rec. 1979. 105:328–9.
27). Gould AR., Kattenbelt JA., Selleck P., Hansson E., Della-Porta A., Westbury HA. Virulent Newcastle disease in Australia: molecular epidemiological analysis of viruses isolated prior to and during the outbreaks of 1998-2000. Virus Res. 2001. 77:51–60.

28). Hanson BA., Swayne DE., Senne DA., Lobpries DS., Hurst J., Stallknecht DE. Avian influenza viruses and paramyxoviruses in wintering and resident ducks in Texas. J Wildl Dis. 2005. 41:624–8.

29). Kawamura M., Nagata-Matsubara K., Nerome K., Yamane N., Kida H., Kodama H., Izawa H., Mikami T. Antigenic variation of Newcastle disease viruses isolated from wild ducks in Japan. Microbiol Immunol. 1987. 31:831–5.
30). Peroulis I., O'Riley K. Detection of avian paramyxoviruses and influenza viruses amongst wild bird populations in Victoria. Aust Vet J. 2004. 82:79–82.

31). Stanislawek WL., Wilks CR., Meers J., Horner GW., Alexander DJ., Manvell RJ., Kattenbelt JA., Gould AR. Avian paramyxo-viruses and influenza viruses isolated from mallard ducks (Anas platyrhynchos) in New Zealand. Arch Virol. 2002. 147:1287–302.
32). Takakuwa H., Ito T., Takada A., Okazaki K., Kisa H. Potentially virulent Newcastle disease viruses are maintained in migratory waterfowl populations. Jpn J Vet Res. 1998. 45:207–15.
33). Vickers ML., Hanson RP. Characterization of isolates of Newcastle disease virus from migratory birds and turkeys. Avian Dis. 1982. 26:127–33.

34). Jang HJ., Jung BY., Kim ST., Kwon KB., Rho SM., Park CK. Nationalwide seroprevalance of Newcastle disease for recent 5 years in Korea. Kor J Vet Publ Hlth. 2007. 31:249–255.
Go to : 
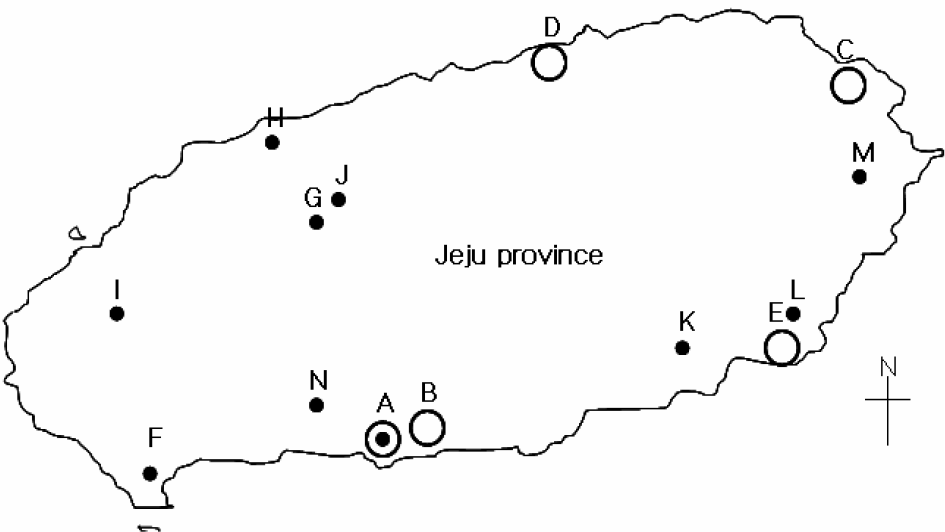 | Figure 1.Geographical distribution of poultry farms where NDV was isolated during 2007~2008. ○, NDV positive farms during first NDV surveillance (2007. 11~2008. 1); •, NDV positive farms during second NDV surveillance (2008. 1~2008. 11) |
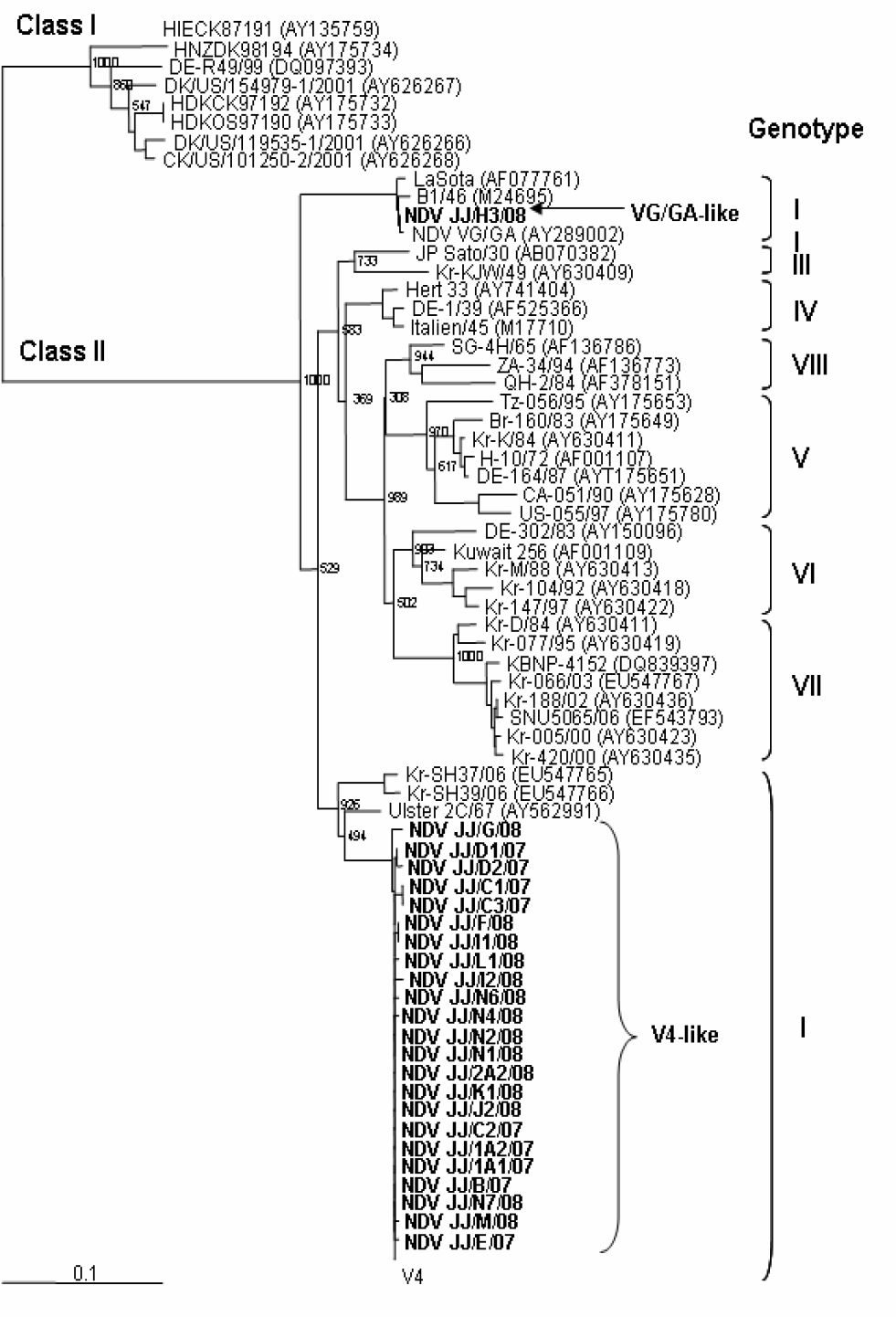 | Figure 2.Phylogenetic analysis of NDV isolates based on the first 389 nucleotides of the coding region of the F gene. Sequences of reference strains were taken from the GenBank database. The accession numbers of reference strains were included in parenthesis. NDV isolates in bold represent NDV isolated in Jeju Province, Korea during 2007~2008 surveillance. |
Table 1.
Lists of farms and their domestic birds sampled in Jeju Province, Korea during 2007~2008 NDV surveillance
| Period sampled | Sample type | Total | Species testeda | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Ly | Br | BB | KN | Dk | Pt | Others | |||
| 1st (‘07.11 ~ ‘08.1) | Swab | 2,506 (63) | 1,520b (36) | 286 (7) | 80 (1) | 400 (14) | 120 (3) | 80 (1) | 20 (1) |
| Faeces | 165 (10) | – | – | – | 45 (2) | 40 (2) | 45 (3) | 35 (3) | |
| 2nd (‘08.8 ~ ‘08.11.) | Swab | 3,054 (68) | 1,798 (31) | 559 (14) | 80 (1) | 597 (21) | 20 (1) | – | – |
| Faeces | 700 (19) | – | – | – | 100 (3) | 270 (7) | 240 (5) | 90 (4) | |
| Total | 6,405 | 3,318 | 845 | 160 | 1,142 | 450 | 365 | 145 | |
Table 2.
History of poultry farms where hemagglutinating agents were isolated in Jeju Province, Korea during 2007~2008 NDV surveillance
| Virusa | Periodb | Flock history | |||
|---|---|---|---|---|---|
| Farm (location) | Bird type | Age (days old) | Vaccination | ||
| JJ/1A1/07 | 1st | A (Seogwipo) | Korean native | 180 | Yes |
| JJ/1A2/07 | 1st | A | Korean native | 210 | Yes |
| JJ/2A2/08c | 2nd | A | Korean native | 280 | No |
| JJ/B1/07 | 1st | B (Seogwipo) | Pheasant | – | Yes |
| JJ/C1/07 | 1st | C (Gujwa) | Duck | 28 | No |
| JJ/C2/07 | 1st | C | Duck | 35 | No |
| JJ/C3/07 | 1st | C | Duck | 21 | No |
| JJ/D1/07 | 1st | D (Jocheon) | Duck | 14 | No |
| JJ/D2/07 | 1st | D | Duck | 56 | Yes |
| JJ/E/07 | 1st | E (Pyoseon) | Korean native | 730 | Yes |
| JJ/F/08 | 2nd | F (Hallim) | Layer | 190 | Yes |
| JJ/G2/08 | 2nd | G (Aewol) | Duck | 35 | Yes |
| JJ/H3/08 | 2nd | H (Aewol) | Korean native | 168 | Yes |
| JJ/I1/08 | 2nd | I (Hangyeong) | Duck | 3 | Yes |
| JJ/I2/08 | 2nd | I | Duck | 21 | Yes |
| JJ/J2/08 | 2nd | J (Aewol) | Korean native | 105 | Yes |
| JJ/K1/08 | 2nd | K (Namwon) | Broiler | 34 | Yes |
| JJ/L1/08 | 2nd | L (Pyoseon) | Broiler | 25 | Yes |
| JJ/M/08 | 2nd | M (Seonsan) | Korean native | 100 | Yes |
| JJ/N1/08 | 2nd | N (Seogipo) | Pheasant | – | Yes |
| JJ/N2/08 | 2nd | N | Pheasant | – | Yes |
| JJ/N4/08 | 2nd | N | Pheasant | – | Yes |
| JJ/N6/08 | 2nd | N | Pheasant | – | Yes |
| JJ/N7/08 | 2nd | N | Pheasant | – | Yes |
Table 3.
Genetic and biological characteristics of NDVs isolated in Jeju Province, Korea during 2007~2008 NDV surveillance
| NDV isolate (Farm) | NDV RT-PCRa | Genotypingb | F0 motif sequence | Heat treatment | Mean death time | |||
|---|---|---|---|---|---|---|---|---|
| Common | Pathotype | Class | Genotype | Before | After | |||
| JJ/1A1/07 (A) | Positive | Negative | I | I | GKQGRLIG | 512 | 64c | >120 |
| JJ/1A2/07 (A) | Positive | Negative | I | I | GKQGRLIG | 512 | 64 | >120 |
| JJ/2A2/08 (A) | Positive | Negative | I | I | GKQGRLIG | 512 | 128 | >120 |
| JJ/B1/07 (B) | Positive | Negative | I | I | GKQGRLIG | 256 | <4 | >120 |
| JJ/C1/07 (C) | Positive | Negative | I | I | GKQGRLIG | 256 | 128 | >120 |
| JJ/C2/07 (C) | Positive | Negative | I | I | GKQGRLIG | 512 | <4 | >120 |
| JJ/C3/07 (C) | Positive | Negative | I | I | GKQGRLIG | 256 | 128 | >120 |
| JJ/D1/07 (D) | Positive | Negative | I | I | GKQGRLIG | 256 | 32 | >120 |
| JJ/D2/07 (D) | Positive | Negative | I | I | GKQGRLIG | 64 | <4 | >120 |
| JJ/E/07 (E) | Positive | Negative | I | I | GKQGRLIG | 128 | 128 | >120 |
| JJ/F/08 (F) | Positive | Negative | I | I | GKQGRLIG | 64 | <4 | >120 |
| JJ/G2/08 (G) | Positive | Negative | I | I | GKQGRLIG | 512 | 256 | >120 |
| JJ/H3/08 (H) | Positive | Negative | I | II | GRQGRLIG | 128 | <4 | >120 |
| JJ/I1/08 (I) | Positive | Negative | I | I | GKQGRLIG | 1024 | 1024 | >120 |
| JJ/I2/08 (I) | Positive | Negative | I | I | GKQGRLIG | 256 | 256 | >120 |
| JJ/J2/08 (J) | Positive | Negative | I | I | GKQGRLIG | 512 | 256 | >120 |
| JJ/K1/08 (K) | Positive | Negative | I | I | GKQGRLIG | 1024 | 32 | >120 |
| JJ/L1/08 (L) | Positive | Negative | I | I | GKQGRLIG | 512 | 256 | >120 |
| JJ/M/08 (M) | Positive | Negative | I | I | GKQGRLIG | 128 | 128 | >120 |
| JJ/N1/08 (N) | Positive | Negative | I | I | GKQGRLIG | 512 | 512 | >120 |
| JJ/N2/08 (N) | Positive | Negative | I | I | GKQGRLIG | 256 | 256 | >120 |
| JJ/N4/08 (N) | Positive | Negative | I | I | GKQGRLIG | 4096 | 1024 | >120 |
| JJ/N6/08 (N) | Positive | Negative | I | I | GKQGRLIG | 512 | 512 | >120 |
| JJ/N7/08 (N) | Positive | Negative | I | I | GKQGRLIG | 2048 | 1024 | >120 |
Table 4.
Amino acid changes in hyper-variable region of fusion protein of NDV isolates




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download