Abstract
A total of 90 Acinetobacter isolates from freshwater and seawater in Gangjin Bay of Korea was investigated for the distribution of genomic species, antimicrobial resistance patterns and clonal relatedness. By amplified ribosomal DNA restriction analysis, eighty-nine Acinetobacter isolates were classified into 11 Acinetobacter genomic species. A. johnsonii (n=23) was the most prevalent, followed by A. baumannii (n=13), A. calcoaceticus (n=13), Acinetobacter genomic species 11 (n=10), A. phenon 6/ct13TU (n=9), A. junii (n=5), A. venetianus (n=5), Acinetobacter genomic species 17 (n=4), 14BJ (n=3), A. phenon 10/1271 (n=2), Acinetobacter genomic species 3 (n=1), and ungrouped (n=1). The majority of Acinetobacter genomic species were isolated from the site A and B, and some known nosocomial pathogens in the clinical environment were observed among them. Of the 11 antimicrobial drugs tested, several A. johnsonii isolates exhibited high-frequency resistance to a wide variety of antimicrobial agents, including ampicillin-sulbactam, piperacillin, ceftazidime, cefotaxime, and sulfamethoxazole (p < 0.001). Some Acinetobacter genomic species were resistant to currently used antibiotics but all isolates were susceptible to imipenem, amikacin, and tetracycline. Based on the results of antimicrobial resistance pattern and phylogenetic analysis, 23 A. johnsonii isolates were classified into 19 pulsotypes. In conclusion, there was a significant difference in the distribution of Acinetobacter species between freshwater and seawater. Predominance of A. johnsonii strains was probably due to their ability to proliferate in the contaminated aquatic environment originated from local geographic features. Therefore, the waste effluent from animals and humans plays an important role in the distribution of Acinetobacter species in aquatic environment.
REFERENCES
1). Bergogne-Bérézin E., Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996. 9:148–65.
2). Nemec A., De Baere T., Tjernberg I., Vaneechoutte M., van der Reijden TJ., Dijkshoorn L. Acinetobacter ursingii sp. nov. and Acinetobacter schindleri sp. nov., isolated from human clinical specimens. Int J Syst Evol Microbiol. 2001. 51:1891–9.
3). Oh JY., Kim KS., Jeong YW., Cho JW., Park JC., Lee JC. Epidemiological typing and prevalence of integrons in multiresistant Acinetobacter strains. APMIS. 2002. 110:247–52.
4). van Dessel H., Dijkshoorn L., van der Reijden T., Bakker N., Paauw A., van den Broek P., Verhoef J., Brisse S. Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res Microbiol. 2004. 155:105–12.
5). Tjernberg I., Ursing J. Clinical strains of Acinetobacter classified by DNA-DNA hybridization. APMIS. 1989. 97:595–605.
6). Chang HC., Wei YF., Dijkshoorn L., Vaneechoutte M., Tang CT., Chang TC. Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J Clin Microbiol. 2005. 43:1632–9.
7). Vaneechoutte M., Dijkshoorn L., Tjernberg I., Elaichouni A., de Vos P., Claeys G., Verschraegen G. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J Clin Microbiol. 1995. 33:11–5.
8). Ko WC., Lee NY., Su SC., Dijkshoorn L., Vaneechoutte M., Wang LR., Yan JJ., Chang TC. Oligonucleotide array-based identification of species in the Acinetobacter calcoaceticus-A. baumannii complex in isolates from blood cultures and antimicrobial susceptibility testing of the isolates. J Clin Microbiol. 2008. 46:2052–9.
9). Lacroix SJ., Cabelli VJ. Membrane filter method for enumeration of Acinetobacter calcoaceticus from environmental waters. Appl Environ Microbiol. 1982. 43:90–6.
10). Goldstein FW., Labigne-Roussel A., Gerbaud G., Carlier C., Collatz E., Courvalin P. Transferable plasmid-mediated antibiotic resistance in Acinetobacter. Plasmid. 1983. 10:138–47.
11). Towner KJ. Clinical Importance and Antibiotic Resistance of Acinetobacter spp. Proceedings of a symposium held on 4~5 November 1996 at Eilat, Israel. J Med Microbiol. 1997. 46:721–46.
12). Guardabassi L., Dalsgaard A., Olsen JE. Phenotypic characterization and antibiotic resistance of Acinetobacter spp. isolated from aquatic sources. J Appl Microbiol. 1999. 87:659–67.
13). Hendrickx L., Hausner M., Wuertz S. Natural genetic transformation in monoculture Acinetobacter sp. strain BD413 biofilms. Appl Environ Microbiol. 2003. 69:1721–7.
14). Schmidt AS., Bruun MS., Dalsgaard I., Larsen JL. Incidence, distribution, and spread of tetracycline resistance determinants and integron-associated antibiotic resistance genes among motile aeromonads from a fish farming environment. Appl Environ Microbiol. 2001. 67:5675–82.

15). Nielsen KM., van Weerelt MD., Berg TN., Bones AM., Hagler AN., van Elsas JD. Natural transformation and availability of transforming DNA to Acinetobacter calcoaceticus in soil microcosms. Appl Environ Microbiol. 1997. 63:1945–52.
16). Baumann P. Isolation of Acinetobacter from soil and water. J Bacteriol. 1968. 96:39–42.
17). National Committee for Clinical Laboratory Standards: Performance standards for antimicrobial susceptibility testing. Approved standard. 2nd ed.M38-A2.CLSI;Wayne, PA: .,. 2008.
18). Gautom RK. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J Clin Microbiol. 1997. 35:2977–80.
19). Seveno NA., Kallifidas D., Smalla K., van EJD., Collard JM., Karagouni AD., Wellington EMH. Occurrence and reservoirs of antibiotic resistance genes in the environment. Rev Med Microbiol. 2002. 13:15–27.
20). Seifert H., Dijkshoorn L., Gerner-Smidt P., Pelzer N., Tjernberg I., Vaneechoutte M. Distribution of Acinetobacter species on human skin: comparison of phenotypic and genotypic identification methods. J Clin Microbiol. 1997. 35:2819–25.
21). Peleg AY., Seifert H., Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008. 21:538–82.
22). Berlau J., Aucken HM., Houang E., Pitt TL. Isolation of Acinetobacter spp. including A. baumannii from vegetables: implications for hospital-acquired infections. J Hosp Infect. 1999. 42:201–4.
23). Lim YM., Shin KS., Kim J. Distinct antimicrobial resistance patterns and antimicrobial resistance harboring genes according to genomic species of Acinetobacter isolates. J Clin Microbiol. 2007. 45:902–5.
24). Seifert H., Strate A., Schulze A., Pulverer G. Vascular catheter-related bloodstream infection due to Acinetobacter johnsonii (formerly Acinetobacter calcoaceticus var. lwoffi): report of 13 cases. Clin Infect Dis. 1993. 17:632–6.
25). Gales AC., Jones RN., Sader HS. Global assessment of the antimicrobial activity of polymyxin B against 54 731 clinical isolates of Gram-negative bacilli: report from the SENTRY antimicrobial surveillance programme (2001~2004). Clin Microbiol Infect. 2006. 12:315–21.

26). Ko KS., Suh JY., Kwon KT., Jung SI., Park KH., Kang CI., Chung DR., Peck KR., Song JH. High rates of resistance to colistin and polymyxin B in subgroups of Acinetobacter baumannii isolates from Korea. J Antimicrob Chemother. 2007. 60:1163–7.
27). Park JC., Lee JC., Oh JY., Jeong YW., Cho JW., Joo HS., Lee WK., Lee WB. Antibiotic selective pressure for the maintenance of antibiotic resistant genes in coliform bacteria isolated from the aquatic environment. Water Sci Technol. 2003. 47:249–53.

28). Infante B., Grape M., Larsson M., Kristiansson C., Pallecchi L., Rossolini GM., Kronvall G. Acquired sulphonamide resistance genes in faecal Escherichia coli from healthy children in Bolivia and Peru. Int J Antimicrob Agents. 2005. 25:308–12.
29). Toleman MA., Bennett PM., Bennett DM., Jones RN., Walsh TR. Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg Infect Dis. 2007. 13:559–65.
30). Gerner-Smidt P., Frederiksen W. Acinetobacter in Denmark: I. Taxonomy, antibiotic susceptibility, and pathogenicity of 112 clinical strains. APMIS. 1993. 101:815–25.
31). Guardabassi L., Dijkshoorn L., Collard JM., Olsen JE., Dalsgaard A. Distribution and in-vitro transfer of tetracycline resistance determinants in clinical and aquatic Acinetobacter strains. J Med Microbiol. 2000. 49:929–36.
32). Petersen A., Guardabassi L., Dalsgaard A., Olsen JE. Class I integrons containing a dhfrI trimethoprim resistance gene cassette in aquatic Acinetobacter spp. FEMS Microbiol Lett. 2000. 182:73–6.
33). De la Fuente CM., Dauros SP., Bello TH., Domínguez YM., Mella MS., Sepúlveda AM., Zemelman ZR., González RG. Mutations in gyrA and gyrB genes among strains of Gram-negative bacilli isolated from Chilean hospitals and their relation with resistance to fluoroquinolones. Rev Med Chil. 2007. 135:1103–10.
34). Unicomb L., Ferguson J., Riley TV., Collignon P. Fluoroquinolone resistance in Campylobacter absent from isolates, Australia. Emerg Infect Dis. 2003. 9:1482–3.
Figure 1.
Phylogenetic analysis of 23 A. johnsonii isolates from the Gangjin Bay of Korea. CHEF electrophoresis of ApaI-digested genomic DNAs. The dendrogram is based on cluster analysis by the unweighted-pair group method with average linkages.
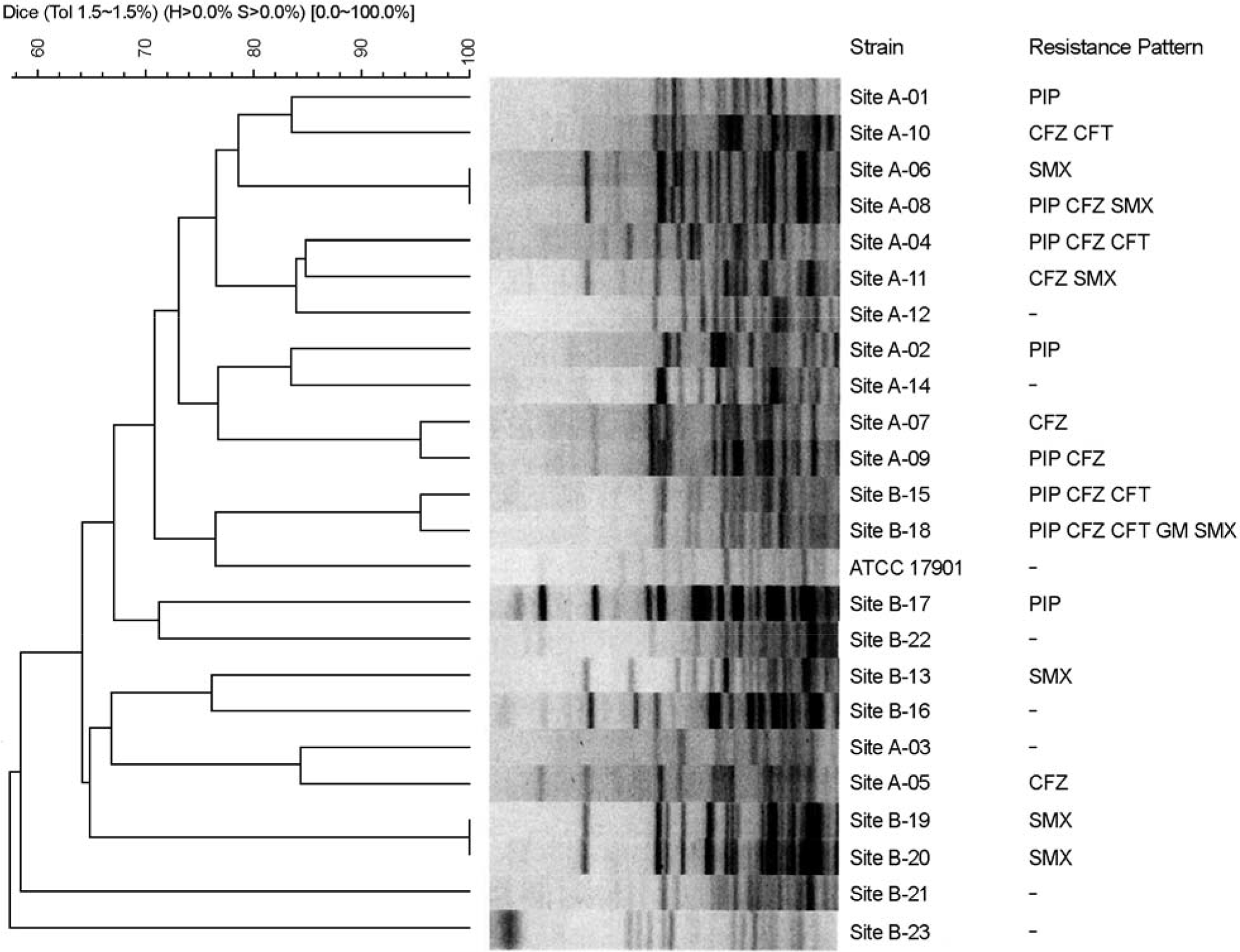
Table 1.
Physicochemical characteristics of sampled water at survey sites
Table 2.
Distribution of Acinetobacter genomic species in Gangjin Bay in Korea
Table 3.
MICs and the percentages of resistance (%R) of 90 Acinetobacter isolates from Gangjin Bay of Korea
| Acinetobacter genomic species (n=90) | MIC90 and %R of a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMSb | PIP | CAZ | CTX | IP | CS | GM | AMK | CIP | TET | SMX | |
| A. johnsonii (n=23) | >128 / 4.4 | ≥512 / 5.6 | 128 / 10.0 | 128 / 4.4 | <1 / 0 | <1 / 0 | <4 / 1.1 | 2 / 0 | 0.5 / 2.2 | 4 / 0 | ≥1,024 / 7.8 |
| A. calcoaceticus (n=13) | 4 / 0 | ≥512 / 2.2 | 4 / 1.1 | 16 / 0 | <1 / 0 | <1 / 0 | <4 / 0 | 4 / 0 | <0.25 / 0 | 2 / 0 | 32 / 0 |
| A. baumannii (n=13) | 4 / 0 | ≥512 / 2.2 | 8 / 0 | 16 / 0 | <1 / 0 | 4 / 0 | <4 / 0 | 8 / 0 | <0.25 / 0 | 4 / 0 | 16 / 0 |
| 11 (n=10) | 2 / 0 | 64 / 1.1 | 8 / 1.1 | 16 / 0 | <1 / 0 | 8 / 5.6 | <4 / 0 | 4 / 0 | 8 / 4.4 | 4 / 0 | 16 / 0 |
| A. phenon 6ct:13TU (n=9) | 4 / 0 | ≥512 / 3.3 | 8 / 0 | 16 / 0 | <1 / 0 | <1 / 0 | <4 / 0 | 4 / 0 | <0.25 / 0 | 2 / 0 | <8 / 0 |
| A. junii (n=6) | <0.5 / 0 | 32 / 1.1 | 4 / 0 | 8 / 0 | 4 / 0 | 8 / 3.3 | <4 / 0 | 4 / 0 | 0.5 / 0 | 4 / 0 | 64 / 1.1 |
| A. venetianus (n=5) | <0.5 / 0 | 16 / 0 | 4 / 0 | 8 / 0 | <1 / 0 | 4 / 0 | <4 / 0 | 8 / 0 | <0.25 / 0 | 2 / 0 | <8 / 0 |
| 17 (n=4) | 2 / 0 | 8 / 0 | <2 / 0 | <2 / 0 | <1 / 0 | 4 / 0 | <4 / 0 | 2 / 0 | <0.25 / 0 | 4 / 0 | 16 / 0 |
| 14BJ (n=3) | 2 / 0 | 32 / 0 | 8 / 0 | 16 / 0 | <1 / 0 | 4 / 0 | <4 / 0 | 4 / 0 | 8 / 1.1 | 2 / 0 | <8 / 0 |
| A. phenon 10/1271 (n=2) | 4 / 0 | 8 / 0 | <2 / 0 | 4 / 0 | <1 / 0 | <1 / 0 | <4 / 0 | 4 / 0 | <0.25 / 0 | 2 / 0 | <8 / 0 |
| 3 (n=1) | 1 / 0 | 32 / 0 | 4 / 0 | 16 / 0 | <1 / 0 | <1 / 0 | <4 / 0 | 2 / 0 | <0.25 / 0 | 2 / 0 | <8 / 0 |
| Ungrouped (n=1) | 4 / 0 | <4 / 0 | <2 / 0 | 4 / 0 | <1 / 0 | 8 / 1.1 | <4 / 0 | 2 / 0 | <0.25 / 0 | 4 / 0 | <8 / 0 |
a AMS, ampicillin-sulbactam (MIC range, 0.5 to 128 μg/ml); PIP, piperacillin (4 to 512 μg/ml); CAZ, ceftazidime (2 to 128 μg/ml); CTX; cefotaxime (2 to 256 μg/ml); CS; colistin (1 to 32 μg/ml); AMK, amikacin (1 to 64 μg/ml); GM, gentamicin (4 to 256 μg/ml); TET, tetracycline (1 to 16 μg/ml); CIP, ciprofloxacin (0.25 to 32 μg/ml); SMX, sulfamethoxazole (8 to 1,024 μg/ml).




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download