Abstract
Hepatitis A virus (HAV) is one of the most important causes of acute infectious hepatitis. The aim of this study was to determine the genotypes of HAV that have been circulating in Koreans. A total of 76 sera referred to our institute for HAV genotyping from 11 Korean provinces were used for this study. Those samples were diagnosed by positive of IgM anti-HAV. HAV RNA was extracted from 150 μl of serum, and reverse transcription PCR-sequencing was used to detect and characterize HAV RNA. Primer pairs from the VP1/2A region of the HAV were used for amplification and sequencing. HAV RNA was found in 64.5% (n = 49) of the 76 patient sera with acute hepatitis A. Forty-seven strains were genotype IIIA in a total of 49 isolated strains (95.9%, 47/49); only two strains belonged to genotype IA (4.1%, 2/49). Thirty eight genotype IIIA isolates were 100% identical to consensus amino acid sequences of the reference strain AJ299467. The amino acid change of L772F was found in two IIIA strains; other IIIA isolates showed one amino acid change. Amino acid of genotype IA was compared to reference strain L20541. K801R was found in 1 strain and Q810S in both strains. The amino acid change of K801R was the first report in Koreans. Until recently HAV genotype IA has been reported as a major circulating HAV genotype in Koreans. In the present study, the predominant HAV strain in Koreans seemed to be HAV genotype IIIA.
Go to : 
REFERENCES
1). De Paula VS., Niel C., Teves SC., Villa LM., Virgolino H., Gaspar AM. Molecular epidemiology of hepatitis A virus in Brazilian Amazon. J Gastroenterol Hepatol. 2006. 21:1435–8.

2). Gust ID. Epidemiological patterns of hepatitis A in different parts of the world. Vaccine. 1992. 10:S56–8.

3). Barzaga BN. Hepatitis A shifting epidemiology in South-East Asia and China. Vaccine. 2000. 18:S61–4.

4). Fix AD., Martin OS., Gallicchio L., Vial PA., Lagos R. Age-specific prevalence of antibodies to hepatitis A in Santiago, Chile: risk factors and shift in age of infection among children and young adults. Am J Trop Med Hyg. 2002. 66:628–32.

5). Sohn YM., Rho HO., Park MS., Park JH., Choi BY., Ki MR., Jang WI. The changing epidemiology of hepatitis A in children and the consideration of active immunization in Korea. Yonsei Med J. 2000. 41:34–9.

6). Costa-Mattioli M., Ferre V., Monpoeho S., Garcia L., Colina R., Billaudel S., Vega I., Perez-Bercoff R., Cristina J. Genetic variability of hepatitis A virus in South America reveals heterogeneity and co-circulation during epidemic outbreaks. J Gen Virol. 2001. 82:2647–52.

7). Robertson BH., Jansen RW., Khanna B., Totsuka A., Nainan OV., Siegl G., Widell A., Margolis HS., Isomura S., Ito K. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J Gen Virol. 1992. 73:1365–77.

8). Fujiwara K., Yokosuka O., Imazeki F., Saisho H., Saotome N., Suzuki K., Okita K., Tanaka E., Omata M. Analysis of the genotype-determining region of hepatitis A viral RNA in relation to disease severities. Hepatol Res. 2003. 25:124–34.

9). De Paula VS., Baptista ML., Lampe E., Niel C., Gaspar AM. Characterization of hepatitis A virus isolates from subgenotypes IA and IB in Rio de Janeiro, Brazil. J Med Virol. 2002. 66:22–7.

10). Chironna M., Grottola A., Lanave C., Villa E., Barbuti S., Quarto M. Genetic analysis of H AV strains recovered from patients with acute hepatitis from Southern Italy. J Med Virol. 2003. 70:343–9.
11). Villar LM., Morais LM., Aloise R., Melo MM., Calado IA., Lampe E., Gaspar AM. Co-circulation of genotypes IA and IB of hepatitis A virus in Northeast Brazil. Braz J Med Biol Res. 2006. 39:873–81.

12). Robertson BH., Khanna B., Nainan OV., Margolis HS. Epidemiologic patterns of wild-type hepatitis A virus determined by genetic variation. J Infect Dis. 1991. 163:286–92.

13). Stene-Johansen K., Skaug K., Blystad H., Grinde B. A unique hepatitis A virus strain caused an epidemic in Norway associated with intravenous drug abuse. The Hepatitis A Study Group. Scand J Infect Dis. 1998. 30:35–8.
14). Heitmann A., Laue T., Schottstedt V., Dotzauer A., Pichl L. Occurrence of hepatitis A virus genotype III in Germany requires the adaptation of commercially available diagnostic test systems. Transfusion. 2005. 45:1097–105.

15). Davidkin I., Zheleznova N., Jokinen S., Gorchakova O., Broman M., Mukomolov S. Molecular epidemiology of hepatitis A in St. Petersburg, Russia, 1997~2003. J Med Virol. 2007. 79:657–62.

16). Kim JS., Kim SH. Molecular epidemiology of an outbreak of hepatitis A in Korea. Korean J Clin Pathol. 2001. 21:114–8.
17). Park JY., Lee JB., Jeong SY., Lee SH., Lee MA., Choi HJ. Molecular characterization of an acute hepatitis A outbreak among healthcare workers at a Korean hospital. J Hosp Infect. 2007. 67:175–81.

18). Park SH., Byun KS., Song JW., Kim JH., Song KJ., Baek LJ., Kwon OS., Yeon JE., Kim JS., Pak YT., Lee CH. Molecular epidemiology of Korean strains of hepatitis A virus. Korean J Hepatol. 2000. 6:276–86.
19). Yun H., Kim S., Lee H., Byun KS., Kwon SY., Yim HJ., Lim YS., Jeong SH., Jee Y. Genetic analysis of HAV strains isolated from patients with acute hepatitis in Korea, 2005~2006. J Med Virol. 2008. 80:777–84.

20). Takahashi H., Yotsuyanagi H., Yasuda K., Koibuchi T., Suzuki M., Kato T., Nakamura T., Iwamoto A., Nishioka K., Iino S., Koike K., Itoh F. Molecular epidemiology of hepatitis A virus in metropolitan areas in Japan. J Gastroenterol. 2006. 41:981–6.

21). Thompson JD., Gibson TJ., Plewniak F., Jeanmougin F., Higgings DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997. 25:4876–82.

22). Jansen RW., Siegl G., Lemon SM. Molecular epidemiology of human hepatitis A virus defined by an antigen-capture polymerase chain reaction method. Proc Natl Acad Sci USA. 1990. 87:2867–71.

23). Pina S., Buti M., Jardi R., Clemente-Casares P., Jofre J., Girones R. Genetic analysis of hepatitis A virus strains recovered from the environment and from patients with acute hepatitis. J Gen Virol. 2001. 82:2955–63.

24). Wattanasri N., Ruchusatsawat K., Wattanasri S. Phylogenetic analysis of hepatitis A virus in Thailand. J Med Virol. 2005. 75:1–7.

25). Costa-Mattioli M., Cristina J., Romero H., Perez-Bercof R., Casane D., Colina R., Garcia L., Vega I., Glikman G., Romanowsky V., Castello A., Nicand E., Gassin M., Billaudel S., Ferre V. Molecular evolution of hepatitis A virus: a new classification based on the complete VP1 protein. J Virol. 2002. 76:9516–25.

26). Byun KS., Kim JH., Song KJ., Baek LJ., Song JW., Park SH., Kwon OS., Yeon JE., Kim JS., Bak YT., Lee CH. Molecular epidemiology of hepatitis A virus in Korea. J Gastroenterol Hepatol. 2001. 16:519–24.
Go to : 
 | Figure 1.Neighbor-joining phylogenetic tree nucleotide sequences in the VP1/P2A region of H AV isolates in Korea. The tree includes reference strains L20541, L07676, L20553, L07722 and L07717 in genotype IA, L07703 in genotype IB, L07729 in genotype IIB, L07725, L0766 and AJ2994678 in genotype IIIA, L205328 in genotype IIIB, L077328 in genotype IV, L07731 in genotype VI. The horizontal bar at the bottom represents a genetic distance of 0.02. |
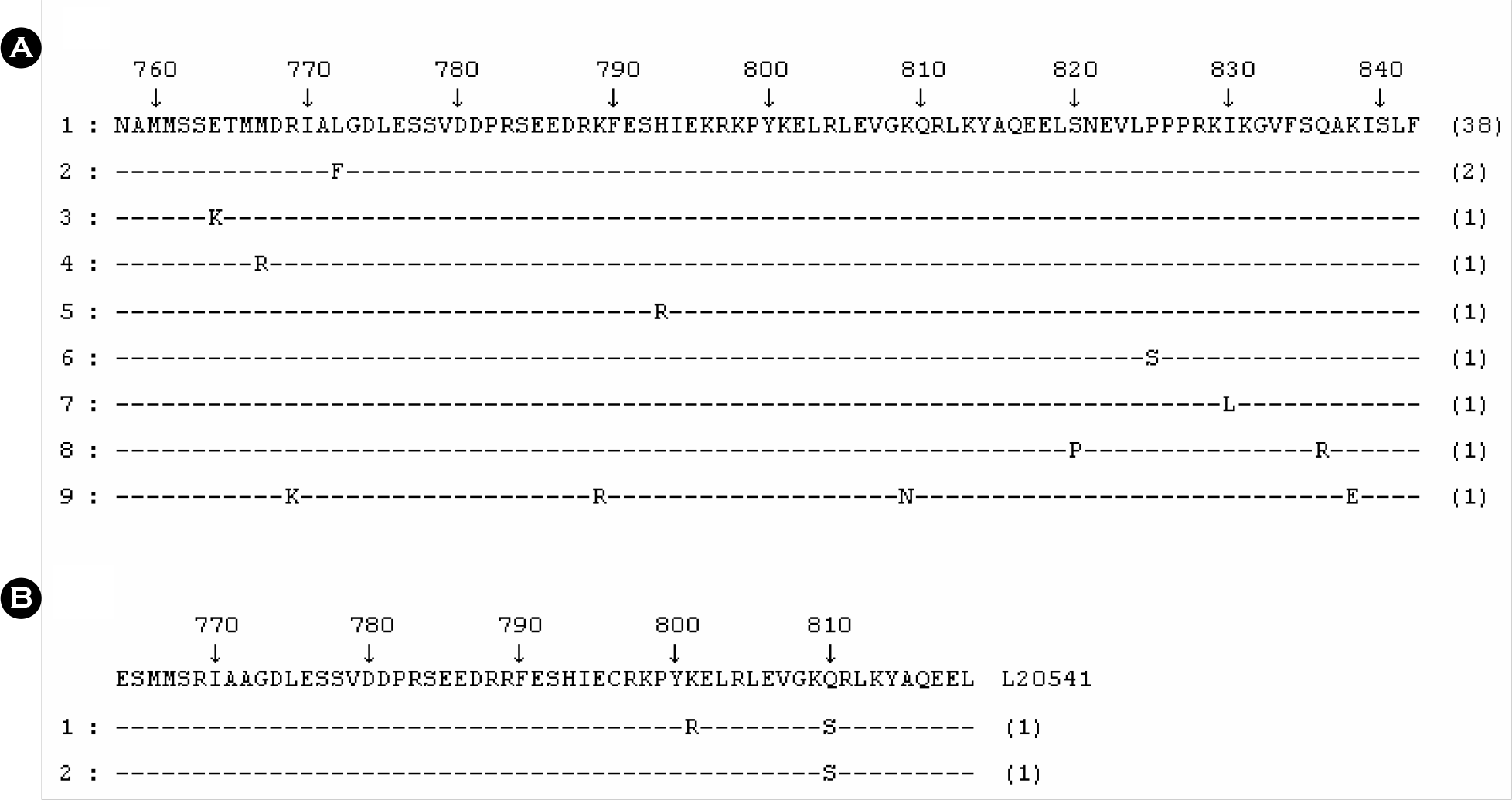 | Figure 2.Comparison of the predicted amino acid sequences of the VP1/VP2 junction region of HAV. The consensus amino acid sequences of AJ299467 for HAV genotype IIIA and L20541 for HAV genotype IA are shown on the top line of (A) and (B), respectively. “–” indicates conserved amino acid; differences are shown by the appropriate single letter amino acid code. The numbers above the consensus amino acid sequence indicate the predicted amino acid number from the start of HAV full amino acid. The numbers of the right ends indicate the number of HAV strains we found in the present study. |
Table 1.
Molecular characters and nucleic acid sequences homologies of 49 strains at the VP1/2A region of hepatitis A Virus (HAV) in Korean HAV isolates compared to reference strains, AJ299467 and L20541 for genotype IIIA and IA, respectively
Table 2.
Amino acid changes of Korean HAV strains in VP1/2A region of HAV from 2007 to 2008




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download