1). Jian JW., Lai CT., Kuo CY., Kuo SH., Hsu LC., Chen PJ., Wu HS., Liu MT. Genetic analysis and evaluation of the reassortment of influenza B viruses isolated in Taiwan during the 2004~2005 and 2006~2007 epidemics. Virus Res. 2008. 131:243–9.

2). Wright PF., Webster RG. Orthomyxoviruses. Fields K.D.M., Hawley B.N., editorsEditors,. Virology. Lippencott-Raven;Philadelphia: 2001. pp.p. 1533–79.
3). Mitnaul LJ., Matrosovich MN., Castrucci MR., Tuzikov AB., Bovin NV., Kobasa D., Kawaoka Y. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J Virol. 2000. 74:6015–20.

4). Rota PA., Wallis TR., Harmon MW., Rota JS., Kendal AP., Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990. 175:59–68.

5). Xu X., Lindstrom SE., Shaw MW., Smith CB., Hall HE., Mungall BA., Subbarao K., Cox NJ., Klimov A. Reassortment and evolution of current human influenza A and B viruses. Virus Res. 2004. 103:55–60.

6). Betakova T., Nermut MV., Hay AJ. The NB protein is an integral component of the membrane of influenza B virus. J Gen Virol. 1996. 77:2689–94.

7). Brassard DL., Leser GP., Lamb RA. Influenza B virus NB glycoprotein is a component of the virion. Virology. 1996. 220:350–60.

8). Briedis DJ., Tobin M. Influenza B virus genome: complete nucleotide sequence of the influenza B/Lee/40 virus genome RNA segment 5 encoding the nucleoprotein and comparison with the B/Singapore/222/79 nucleoprotein. Virology. 1984. 133:448–55.

9). Briedis DJ., Lamb RA., Choppin PW. Sequence of RNA segment 7 of the influenza B virus genome: partial amino acid homology between the membrane proteins (M1) of influenza A and B viruses and conservation of a second open reading frame. Virology. 1982. 116:581–8.

10). Kanegae Y., Sugita S., Endo A., Ishida M., Senya S., Osako K., Nerome K., Oya A. Evolutionary pattern of the hemagglutinin gene of influenza B viruses isolated in Japan: cocirculating lineages in the same epidemic season. J Virol. 1990. 64:2860–5.

11). Rota PA., Hemphill ML., Whistler T., Regnery HL., Kendal AP. Antigenic and genetic characterization of the haemagglutinins of recent cocirculating strains of influenza B virus. J Gen Virol. 1992. 73:2737–42.

12). Air GM., Gibbs AJ., Laver WG., Webster RG. Evolutionary changes in influenza B are not primarily governed by antibody selection. Proc Natl Acad Sci USA. 1990. 87:3884–8.

13). Luo C., Morishita T., Satou K., Tateno Y., Nakajima K., Nobusawa E. Evolutionary pattern of influenza B viruses based on the HA and NS genes during 1940 to 1999: origin of the NS genes after 1997. Arch Virol. 1999. 144:1881–91.

14). McCullers JA., Wang GC., He S., Webster RG. Reassortment and insertion-deletion are strategies for the evolution of influenza B viruses in nature. J Virol. 1999. 73:7343–8.

15). Nerome R., Hiromoto Y., Sugita S., Tanabe N., Ishida M., Matsumoto M., Lindstrom SE., Takahashi T., Nerome K. Evolutionary characteristics of influenza B virus since its first isolation in 1940: dynamic circulation of deletion and insertion mechanism. Arch Virol. 1998. 143:1569–83.

16). Barr IG., Komadina N., Hurt A., Shaw R., Durrant C., Iannello P., Tomasov C., Sjogren H., Hampson AW. Reassortants in recent human influenza A and B isolates from South East Asia and Oceania. Virus Res. 2003. 98:35–44.

17). Chen GW., Shih SR., Hsiao MR., Chang SC., Lin SH., Sun CF., Tsao KC. Multiple genotypes of influenza B viruses cocirculated in Taiwan in 2004 and 2005. J Clin Microbiol. 2007. 45:1515–22.

18). Nah SY., Park SE., Park JY., Lee HJ. Epidemiology of influenza virus over 8 years (1990~1998) in Seoul, Korea. Korean J Infect Dis. 1999. 31:210–6.
19). KCDC. Influenza Sentinel Surveillance Report, 2007~2008. Number 29. 2008.
20). Yamashita M., Krystal M., Fitch WM., Palese P. Influenza B virus evolution: co-circulating lineages and comparison of evolutionary pattern with those of influenza A and C viruses. Virology. 1988. 163:112–22.

21). Pechirra P., Nunes B., Coelho A., Ribeiro C., Gonçalves P., Pedro S., Castro LC., Rebelo-de-Andrade H. Molecular characterization of the HA gene of influenza type B viruses. J Med Virol. 2005. 77:541–9.

22). Lindstrom SE., Hiromoto Y., Nishimura H., Saito T., Nerome R., Nerome K. Comparative analysis of evolutionary mechanisms of the hemagglutinin and three internal protein genes of influenza B virus: multiple cocirculating lineages and frequent reassortment of the NP, M, and NS genes. J Virol. 1999. 73:4413–26.

23). Hwang YO., Seo BT., Choi BY. Analysis of isolation and subtyping of influenza virus in Seoul, during 1999~2003. J Bacteriol Virol. 2004. 34:67–74.
24). Hoffmann E., Mahmood K., Yang CF., Webster RG., Greenberg HB., Kemble G. Rescue of influenza B virus from eight plasmids. Proc Natl Acad Sci USA. 2002. 99:11411–6.

25). Stockton J., Ellis JS., Saville M., Clewley JP., Zambon MC. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J Clin Microbiol. 1998. 36:2990–5.

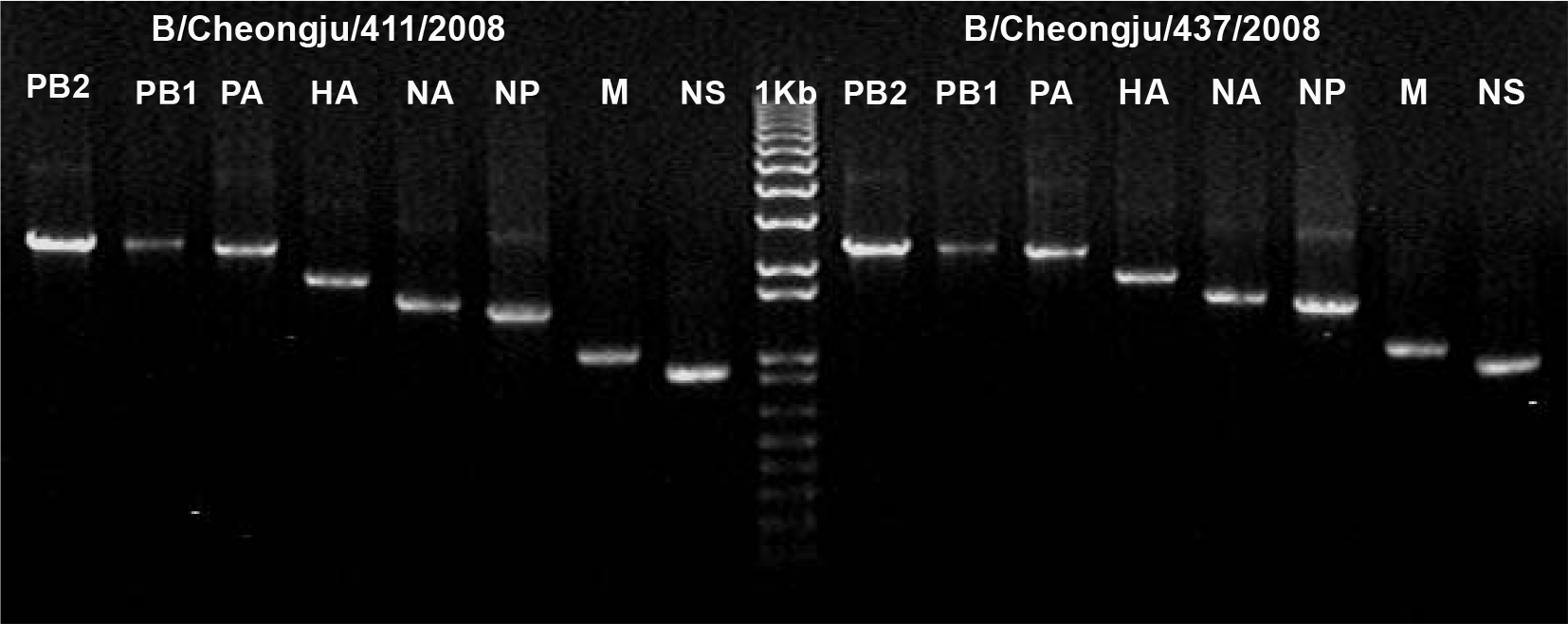
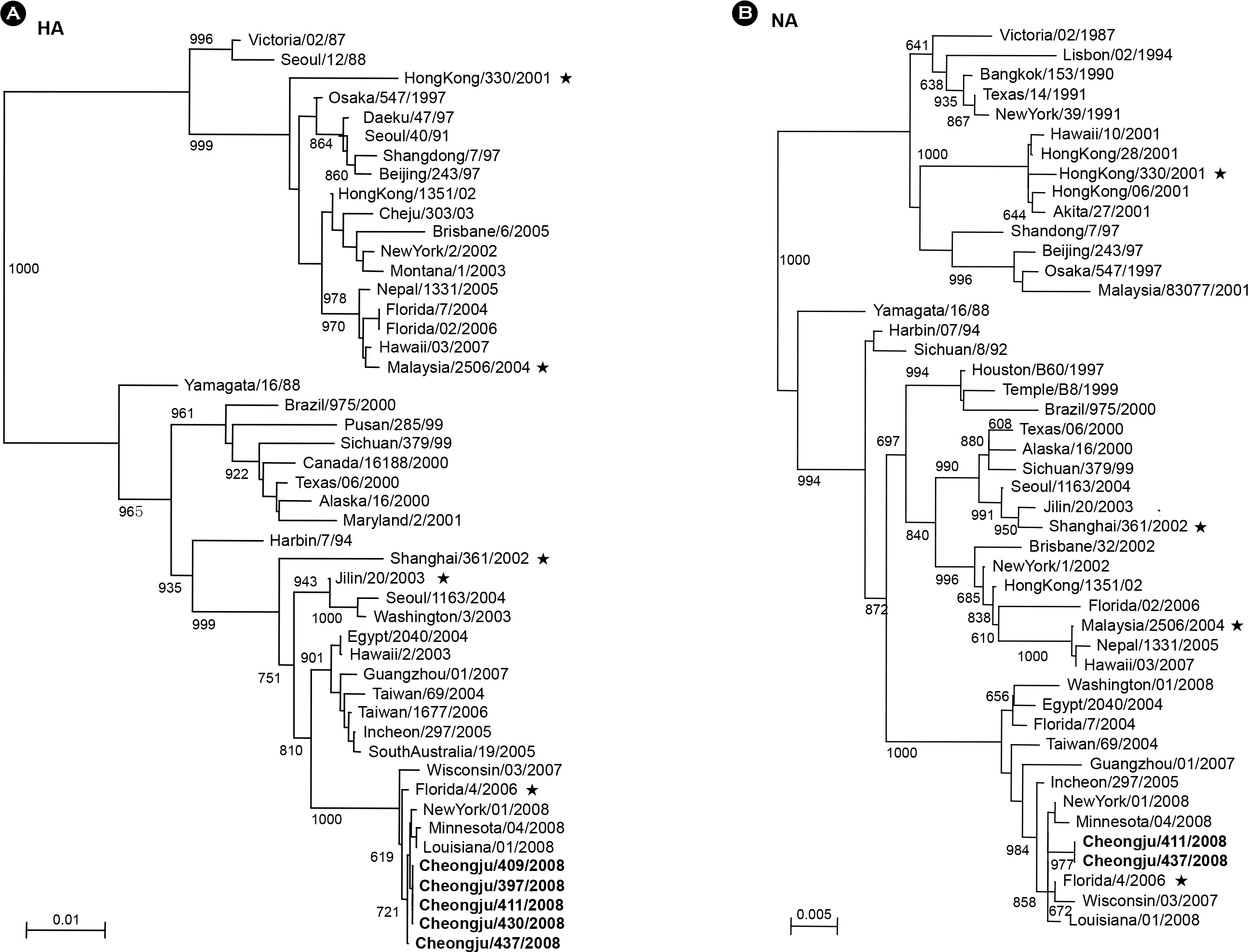
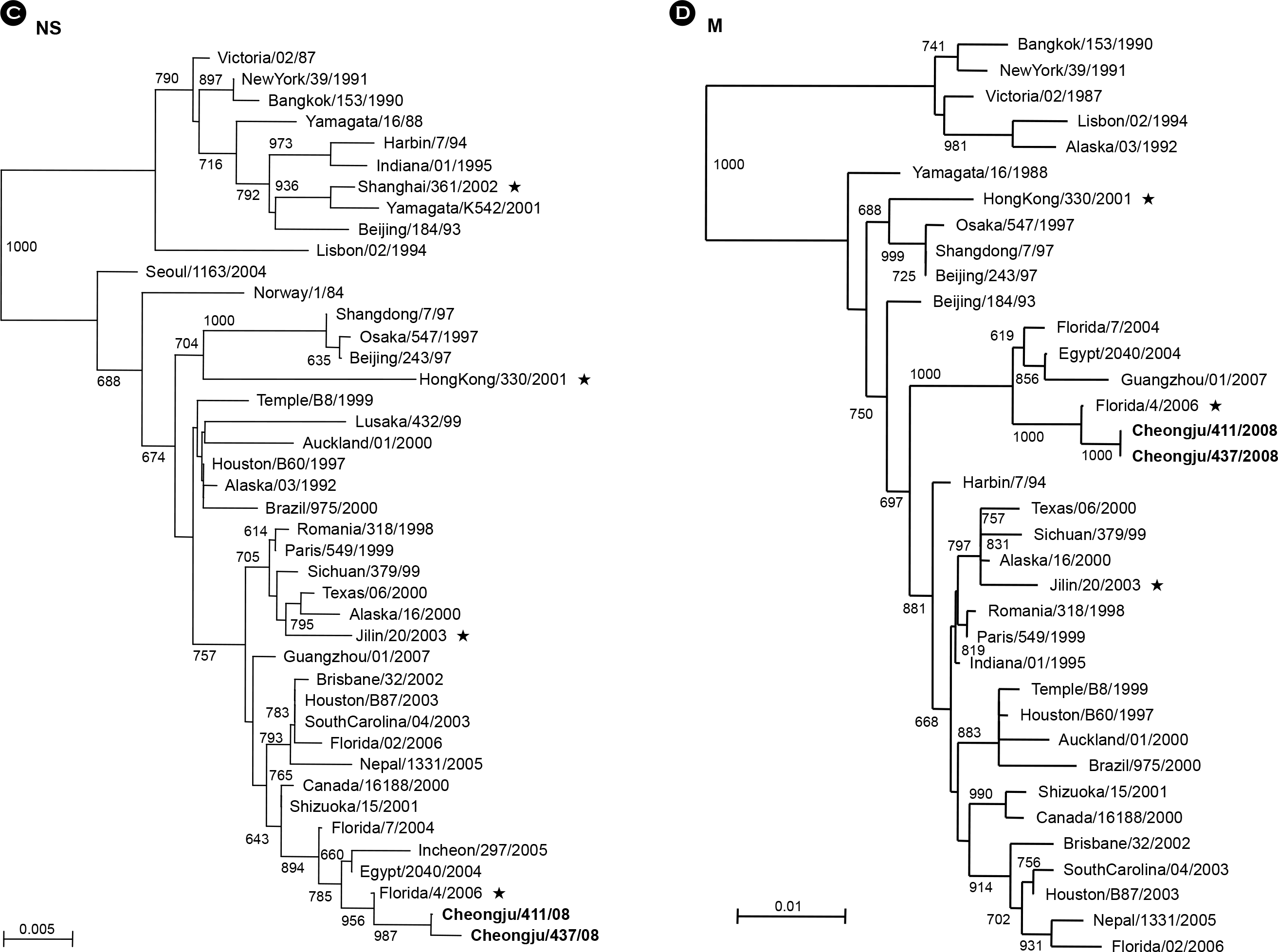
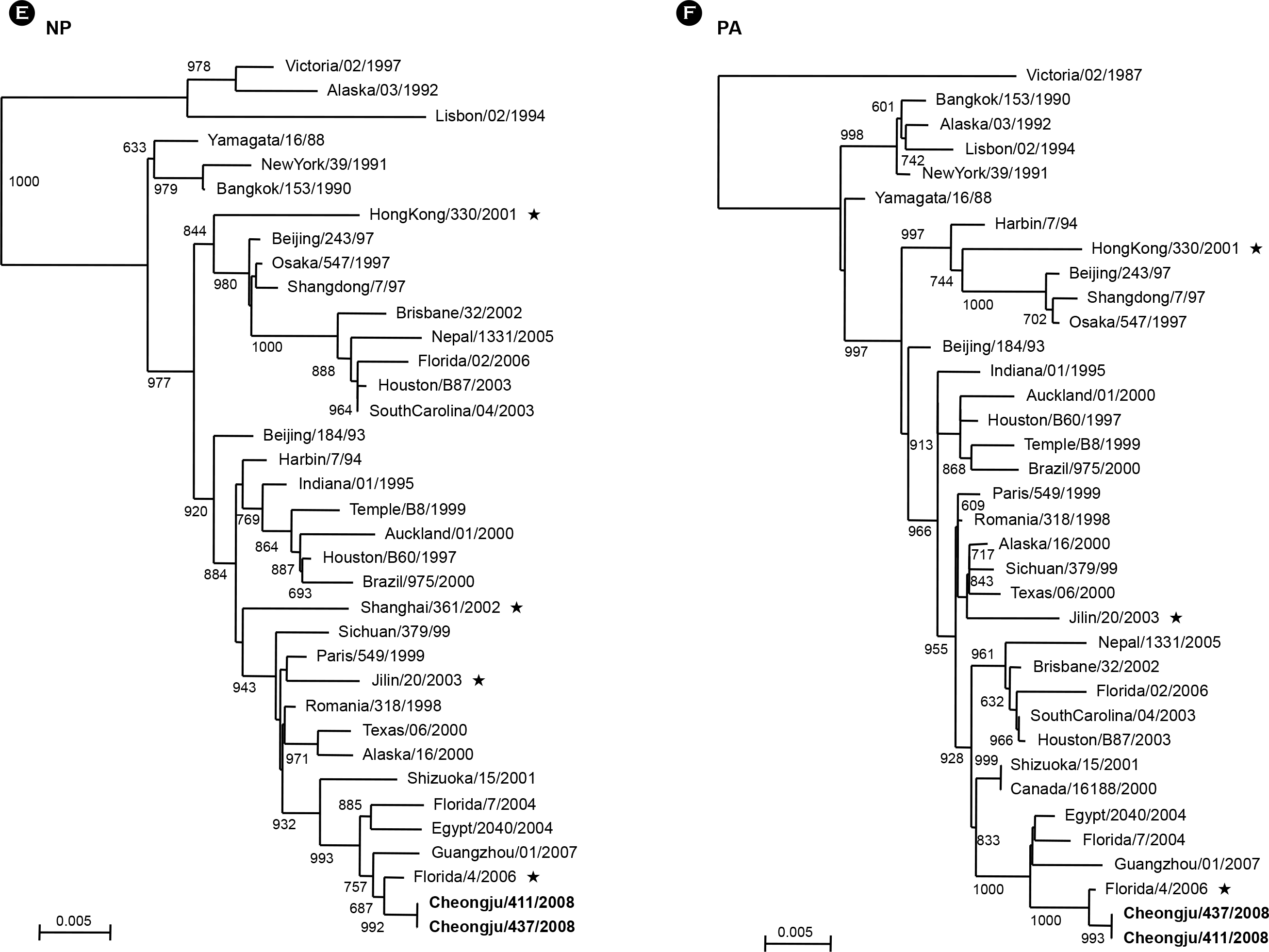




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download