Abstract
The cytolysin A (ClyA) is a 34 kDa pore-forming cytotoxic protein and expressed by some enteric bacteria including Salmonella typhi. This toxin is transported on the bacterial surface and secreted without posttranslational modification. Using the surface display of ClyA, the expression vectors for 193-aa immunogenic antigen of spike protein (termed S1E) from severe acute respiratory syndrome coronavirus (SARS-CoV) were constructed. The vectors carried a gene encoding S. typhi ClyA conjugated to S1E at the C terminus (termed ClyA-S1E) and asd gene in pGEM-T and pBR322, named pGApLCS1E and pBApLCS1E, respectively. An asd-mutated E. coli transformed with these vectors could grow without diaminopimelic acid (DAP), indicating that they were stably maintained in such mutants. ClyA-S1E recombinant proteins from these vectors were expressed on the surface of the attenuated S. typhimurium deficient of global virulence gene regulator, ppGpp. However, they did not show the hemolytic activity on the blood agar plate and cytotoxicity against HeLa cells. To examine whether bacteria expressing ClyA-S1E induced the immune response against S1E, S. typhimurium deficient of ppGpp and Asd was transformed with these vectors and orally immunized in mice. In the western blotting against GST-conjugated S1E using the immunized mouse sera, it was shown that the significant band was detected in the mouse serum by the bacteria transformed with pGApLCS1E but not with pBApLCS1E. It indicates that the immune response producing antibody was dependent on the expression level of ClyA-S1E. Therefore, ClyA delivery system can be used for SARS vaccine development.
REFERENCES
1). Garmory HS., Brown KA., Titball RW. Salmonella vaccines for use in humans: present and future perspectives. FEMS Microbiol Rev. 2002. 26:339–53.
2). Garmory HS., Leary SE., Griffin KF., Williamson ED., Brown KA., Titball RW. The use of live attenuated bacteria as a delivery system for heterologous antigens. J Drug Target. 2003. 11:471–9.

3). Kochi SK., Killeen KP., Ryan US. Advances in the development of bacterial vector technology. Expert Rev Vaccines. 2003. 2:31–43.
4). Lee JS., Shin KS., Pan JG., Kim CJ. Surface-displayed viral antigens on Salmonella carrier vaccine. Nat Biotechnol. 2000. 18:645–8.
5). Gentschev I., Dietrich G., Goebel W. The E. coli alpha-hemolysin secretion system and its use in vaccine development. Trends Microbiol. 2002. 10:39–45.
6). Zhu C., Ruiz-Perez F., Yang Z., Mao Y., Hackethal VL., Greco KM., Choy W., Davis K., Butterton JR., Boedeker EC. Delivery of heterologous protein antigens via hemolysin or autotransporter systems by an attenuated ler mutant of rabbit enteropathogenic Escherichia coli. Vaccine. 2006. 24:3821–31.
7). Wallace AJ., Stillman TJ., Atkins A., Jamieson SJ., Bullough PA., Green J., Artymiuk PJ. E. coli hemolysin E (HlyE, ClyA, SheA): X-ray crystal structure of the toxin and observation of membrane pores by electron microscopy. Cell. 2000. 100:265–76.
8). Wai SN., Lindmark B., Söderblom T., Takade A., Westermark M., Oscarsson J., Jass J., Richter-Dahlfors A., Mizunoe Y., Uhlin BE. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell. 2003. 115:25–35.

9). del Castillo FJ., Moreno F., del Castillo I. Secretion of the Escherichia coli K-12 SheA hemolysin is independent of its cytolytic activity. FEMS Microbiol Lett. 2001. 204:281–5.
10). Galen JE., Zhao L., Chinchilla M., Wang JY., Pasetti MF., Green J., Levine MM. Adaptation of the endogenous Salmonella enterica serovar Typhi clyA-encoded hemolysin for antigen export enhances the immunogenicity of anthrax protective antigen domain 4 expressed by the attenuated live-vector vaccine strain CVD 908-htrA. Infect Immun. 2004. 72:7096–106.
11). Spiga O., Bernini A., Ciutti A., Chiellini S., Menciassi N., Finetti F., Causarono V., Anselmi F., Prischi F., Niccolai N. Molecular modelling of S1 and S2 subunits of SARS coronavirus spike glycoprotein. Biochem Biophys Res Commun. 2003. 310:78–83.

12). Li W., Moore MJ., Vasilieva N., Sui J., Wong SK., Berne MA., Somasundaran M., Sullivan JL., Luzuriaga K., Greenough TC., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003. 426:450–4.

13). Wong SK., Li W., Moore MJ., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004. 279:3197–201.

14). He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M., Jiang S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun. 2004. 324:773–81.

15). Valle E., Guiney DG. Characterization of Salmonella-induced cell death in human macrophage-like THP-1 cells. Infect Immun. 2005. 73:2835–40.
16). Kang HY., Srinivasan J., Curtiss R 3rd. Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar typhimurium vaccine. Infect Immun. 2002. 70:1739–49.
17). Song M., Kim HJ., Kim EY., Shin M., Lee HC., Hong Y., Rhee JH., Yoon H., Ryu S., Lim S., Choy HE. ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J Biol Chem. 2004. 279:34183–90.
18). Galan JE., Nakayama K., Curtiss R 3rd. Cloning and characterization of the asd gene of Salmonella typhimurium: use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene. 1990. 94:29–35.
19). Nayak AR., Tinge SA., Tart RC., McDaniel LS., Briles DE., Curtiss R 3rd. A live recombinant avirulent oral Salmonella vaccine expressing pneumococcal surface protein A induces protective responses against Streptococcus pneumoniae. Infect Immun. 1998. 66:3744–51.
20). Na HS., Kim HJ., Lee HC., Hong Y., Rhee JH., Choy HE. Immune response induced by Salmonella typhimurium defective in ppGpp synthesis. Vaccine. 2006. 24:2027–34.
21). Westermark M., Oscarsson J., Mizunoe Y., Urbonaviciene J., Uhlin BE. Silencing and activation of ClyA cytotoxin expression in Escherichia coli. J Bacteriol. 2000. 182:6347–57.
22). Morales C., Lee MD., Hofacre C., Maurer JJ. Detection of a novel virulence gene and a Salmonella virulence homologue among Escherichia coli isolated from broiler chickens. Foodborne Pathog Dis. 2004. 1:160–5.
23). Lai XH., Arencibia I., Johansson A., Wai SN., Oscarsson J., Kalfas S., Sundqvist KG., Mizunoe Y., Sjöstedt A., Uhlin BE. Cytocidal and apoptotic effects of the ClyA protein from Escherichia coli on primary and cultured monocytes and macrophages. Infect Immun. 2000. 68:4363–7.
Figure 1.
Construction of plasmids. pGApLCS1E and pBApLCS1E were constructed from pGEM-T and pBR322, respectively and carried both of clyA-S1E and asd genes. The clyA-S1E genes are induced by lac promoters.
Figure 2.
Surface display and secretion of ClyA and ClyA-S1E in S. typhimurium. (A) SHJ 2037 strains transformed with the indicated plasmids were lyzed by boiling and separated in 10% SDS-PAGE. After transferring into a membrane, proteins were stained with rabbit anti-ClyA (left panel) and anti-S1E (right panel) antisera. ClyA (34 kDa) and ClyA-S1E (53 kDa) are marked as arrows. 1, bacteria transformed with pUC19; 2, bacteria transformed with pGCSO; 3, bacteria transformed with pGApLCS1E; 4, bacteria transformed with pBApLCS1E. (B) Surface expression of ClyA and ClyAS1E on S. typhimurium SHJ2037. Bacteria transformed with the indicated plasmids were stained with rabbit anti-ClyA antiserum and FITC-conjugated anti-rabbit IgG (right panels), and then stained with DAPI (left panels). The stained bacteria were observed in fluorescence microscopy. 1, bacteria transformed with pUC19; 2, bacteria transformed with pGCSO; 3, bacteria transformed with pGApLCS1E; 4, bacteria transformed with pBApLCS1E. (C) Secretion of ClyA and ClyA-S1E from S. typhimurium SHJ2037. The proteins in the supernatant from bacteria transformed with the indicated plasmids were precipitated in the cold acetone. After washing, the precipitates were separated on 10% SDS-PAGE. After transferring into a membrane, proteins were stained with rabbit anti-ClyA (left panel) and anti-S1E (right panel) antisera. ClyA (34 kDa) and ClyA-S1E (53 kDa) are marked as arrows. The asterisk indicated the degraded remnants from the secreted ClyA-S1E. 1, bacteria transformed with pUC19; 2, bacteria transformed with pGCSO; 3, bacteria transformed with pGApLCS1E; 4, bacteria transformed with pBApLCS1E.
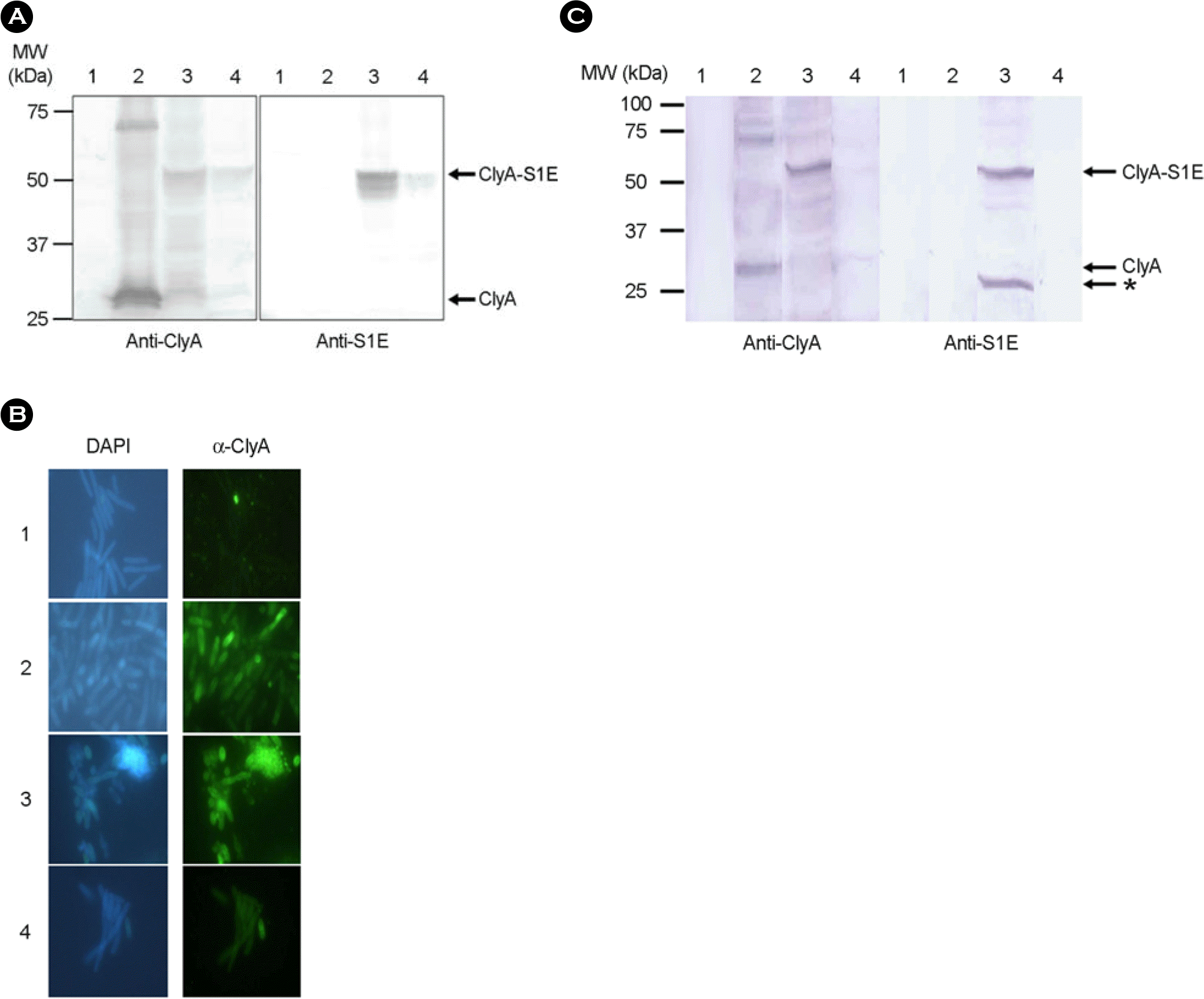
Figure 3.
ClyA-S1E lost hemolytic and cytotoxic acitivities. (A) Hemolysis assay of S. typhimurium strains expressing the ClyA and ClyA-S1E proteins. ppGpp-deficient SHJ2037 (left panels) and hns-mutated SCH2008 with (right panels) transformed with the indicated plasmids were cultivated on the sheep blood agar plate for 2 days. 1, bacteria transformed with pUC19; 2, bacteria transformed with pGCSO; 3, bacteria transformed with pGApLCS1E; 4, bacteria transformed with pBApLCS1E. (B) The cytotoxicity of S. typhimurium expressing ClyA and ClyA-S1E against HeLa cells. The cells (2 × 106) were cultured for 9 hr after inoculation of S. typhimurium SHJ2037 transformed with the indicated plasmids (2 × 108). Cell morphology was observed with microscopy. 1, bacteria transformed with pUC19; 2, bacteria transformed with pGCSO; 3, bacteria transformed with pGApLCS1E; 4, bacteria transformed with pBApLCS1E. (C) The genomic DNA fragmentation by apoptosis was assayed in 2% agarose gel electrophoresis using the same cells in (B).
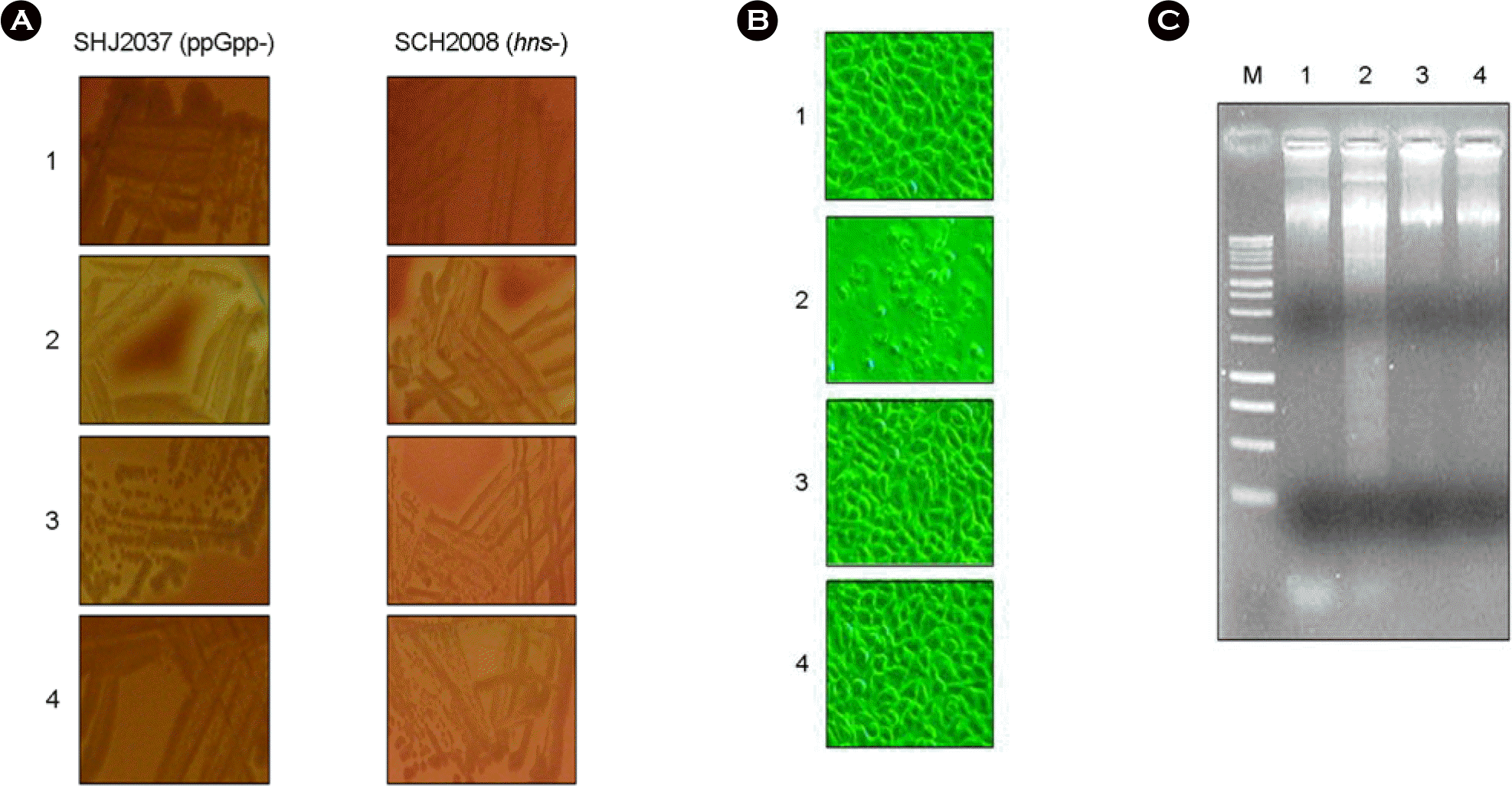
Figure 4.
Detection of anti-S1E antibody in mice orally immunized by S. typhimurium expressing ClyA-S1E. S. typhimurium SHJ2107 strains carrying the deficiency of ppGpp and Asd were transformed with the indicated plasmids. BALB/C mice were orally immunized by the transformed bacteria (5 × 108) and the blood was taken from mice at day 30. The serum was used for western blotting against E. coli lysates expressing GST-conjugated S1E (upper panel). Ponseau S staining of E. coli lysates expressing GST-conjugated S1E separated in SDS-PAGE (lower panel). Western blotting against E. coli lysates containing GST-tagged S1E using the immunized antiserum. As a control, the same sample was western blotted by rabbit anti-S1E antiserum. 1, mouse antiserum after the infection by bacteria with pGA; 2, mouse antiserum after the infection by bacteria with pGApLC; 3, mouse antiserum after the infection by bacteria with pGApLCS1E; 4, mouse antiserum after the infection by bacteria with pBApLCS1E; 5, rabbit anti-S1E antiserum.
Table 1.
Bacterial strains and plasmids used in this study




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download