Abstract
The Bacillus cereus group includes B. anthracis, B. cereus, B. thuringiensis, B. mycoides, B. weihenstephanensis, B. pseudomycoides. The members of B. cereus group shares strong degree of DNA sequence similarity. Even though the biochemical test and bacteriological test have been used to identify the B. cereus group, an accurate identification system of the B. cereus group is required. We have developed a highly specific PCR-based assay for the B. cereus group chromosome using a sequence motif found within a spore structural gene (sspE). Using the assay, we were able to discriminate B. anthracis from the other members of B. cereus group. We also tried to find a new system for the B. cereus group identification. Five bacteriological tests (hemolysis, motility, penicillin susceptibility, rhizoid growth, toxic crystal formation), API system (API 50CHB & API 20E), MLST and sspE PCR were performed on 28 strains of the B. cereus group. The dendrogram generated from API system and bacteriological tests revealed that B. cereus and B. thuringiensis are grouped into the same cluster. In combination of sspE PCR and bacteriological tests, the dendrogram showed that 4 strains of B. cereus clustered within the same group. B. thuringiensis formed the subgroup in the same cluster. All strains of B. mycoides were encompassed together. Another cluster only included B. anthracis. The best system was determined to be sspE PCR and bacteriological tests. It is concluded that sspE PCR and bacteriological tests could be used for rapid discrimination and identification of B. anthracis and provided an effective means of differentiation between the B. cereus group.
References
1). 최철순, 김민희, 정상인, 양용태. 탄저균 병원성 및 약 독백신주의 유사 Bacillus species로부터 간이감별을 위 한 주요 배양 및 생물학적 성상. 대한미생물학회지. 27:93–102. 1992.
2). Anderson GL, Simchock JM, Wilson KH. Identification of a region of genetic variability among Bacillus anthracis strains and related species. J Bacteriol. 178:377–384. 1996.
3). Ash C, Collins MD. Comparative analysis of 23S ribosomal RNA gene sequences of Bacillus anthracis and emetic Bacillus cereus determined by PCR-direct sequencing. FEMS Microbiol Lett. 73:75–80. 1992.
4). Ash C, Farrow JA, Dorsch M, Stackebrandt E, Collins MD. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int J Syst Bacteriol. 41:343–346. 1991.
5). Logan NA, Carman JA, Melling J, Berkeley RC. Identification of Bacillus anthracis by API tests. J Med Microbiol. 20:75–85. 1985.
6). Bourque SN, Valero JR, Lavoie MC, Levesque RC. Comparative Analysis of the 16S to 23S Ribosimal Intergenic Spacer Sequence of Bacillus thuringiensis strains and Subspecies and of Closely Related Species. Appl Environ Microbiol. 61:1623–1626. 1995.
7). Feil EJ. How Stable Are the Core Genes of Bacterial Pathogen? ASM News. 69:234–239. 2003.
8). Gebhardt C, Murray P, Wood WA, Krig NR. Method for general and molecular bacteriology. American Society for Microbiology;Washington, D.C.: 1994.
9). Green BD, Battisti L, Koehler TM, Thorne CB, Ivins BE. Demonstration of capsule plasmid in Bacillus anthracis. Infect Immun. 49:291–297. 1985.
10). Harbottle H, White DG, McDermott PF, Walker RD, Zhao S. Comparison of multilocus sequence typing, pulsed-field gel electrophoresis, and antimicrobial susceptibility typing for characterization of Salmonella enterica serotype newport isolates. J Clin Microbiol. 44:2449–2457. 2006.
11). Helgason E, Tourasse NJ, Meisal R, Caugant DA, Kolst⊘ AB. Multilocus Sequence Typing Scheme for Bacteria of the Bacillus cereus Group. Appl Environ Microbiol. 70:191–201. 2004.
12). Jolley KA, Feil EJ, Chan MS, Maiden MC. Sequence type analysis and recombinational tests (START). Bioinformatics. 17:1230–1231. 2001.

13). Kaneko T, Nozaki R, Aizawa K. Dexoyribonucleic acid relatedness between Bacillus anthracis, Bacillus cereus and Bacillus thuringiensis. Microbiol Immunol. 22:639–641. 1978.
14). Kim K, Cheon E, Wheeler KE, Youn Y, Leighton TJ, Park C, Kim W, Chung S. Derermination of the Most Closely Related Bacillus Isolates to Bacillus anthracis by Multilocus Sequence Typing. Yale J Biol Med. 78:1–14. 2005.
15). Kim KJ, Seo JW, Wheeler K, Park CM, Kim DW, Park SJ, Kim WY, Chung SI, Leighton T. Rapid genotypic detection of Bacillus anthracis and the Bacillus cereus group by multiplex real- time PCR melting curve analysis. FEMS Immunol Microbiol. 43:301–310. 2005.
16). Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics. 17:1244–1245. 2001.

17). Lechner S, Mayr R, Francis KP, Pruss BM, Kaplan T, Wiessner-Gunkel E, Stewart GS, Scherer S. Bacillus weihenstephanensis sp. nov. is a new psychrotolerant species of the Bacillus cereus group. Int J Syst Bacteriol. 48(Pt 4):1373–1382. 1998.
18). Logan NA, Berkeley RCW. Classification and identification fo members of the genus Bacillus using API tests. pp. p. 105–140. In. The Aerobic Endospore-forming Bacteria: Classification and Identification. Berkeley RCW, Goodfellow M, editors. (Ed),. Academic Press;London: 1981.
19). Logan NA, Berkeley RC. Identification fo Bacillus Strains Using the API System. J Gen Microbiol. 130:1871–1882. 1984.
20). Logan NA, Capel BJ, Melling J, Berkeley RC. Distinction between emetic and other strains of Bacillus cereus using the API SYSTEM and Numerical methods. FEMS Microbiology Letters. 5:373–375. 1979.
21). Marston CK, Gee JE, Popovic T, Hoffmaster AR. Molecular approaches to identify and differentiate Bacillus anthracis from phenotypically similar Bacillus species isolates. BMC Microbiol. 6:22. 2006.

22). Morgan U, Ochman H, Renaud F, Tibayrenc M. Population genetics and Population biology: What did they bring to the epidermiology of transmissible disease? Infect Genet Evol. 1:161–166. 2001.
23). Murray PR, Baron EJ. Manual of Clinical Microbiology. Sixth edition. pp. p. 349–356. ASM Press;1995.
24). Nakamura LK. Bacillus pseudomycoides sp. nov. Int J Syst Bacteriol. 48(Pt 3):1031–1035. 1998.
25). Patra G, Sylvestre P, Ramisse V, Thérasse J, Guesdon JL. Isolation of a specific chromosomic DNA sequence of Bacillus anthracis and its possible use in diagnosis. FEMS Immunol Med Microbiol. 15:223–231. 1996.
26). Rhodehamel EJ, Harmon SM. Chapter 14 Bacillus cereus. In. Bacteriological Analytical Manual online. 8th edition.US Department of Health and Human Services and US Food and Drug Administration;2001.
27). Seki T, Chung C, Mikami H, Oshima Y. Deoxyribonucleic acid homology and taxonomy of the genus Bacillus. Int J Syst Bacteriol. 28:182–189. 1978.
28). Sneath PH, Mair NS, Sharpe ME, Holt JG. Bergey's manual of Systematic bacteriology. Vol. 2:pp. p. 1105–1138. Williams & Wilkins;Baltimore, MD: 1986.
29). Turnbull PC, Hutson RA, Ward MJ, Jones MN, Quinn CP, Finnie NJ, Duggleby CJ, Kramer JM, Melling J. Bacillus anthracis but not always anthrax. J Appl Bacteriol. 72:21–28. 1992.
30). Yamada S, Ohashi E, Agata N, Venkateswaran K. Cloning and nucleotide sequence analysis of gyrB of Bacillus cereus, B. thuringiensis, B. mycoides, and B. anthracis and their application to the detection of B. cereus in rice. Appl Environ Microbiol. 65:1483–1490. 1999.
Figure 1.
Dendrogram of 28 strains of B. cereus group constructed by UPGMA based on the bacteriological and API System (API 50CHB & API 20E). The clades are indicated by roman numerals. Clade II encompassed 4 strains of B. anthracis. The scale bars indicate the percentage of missmatches between biological and biochemical properties.
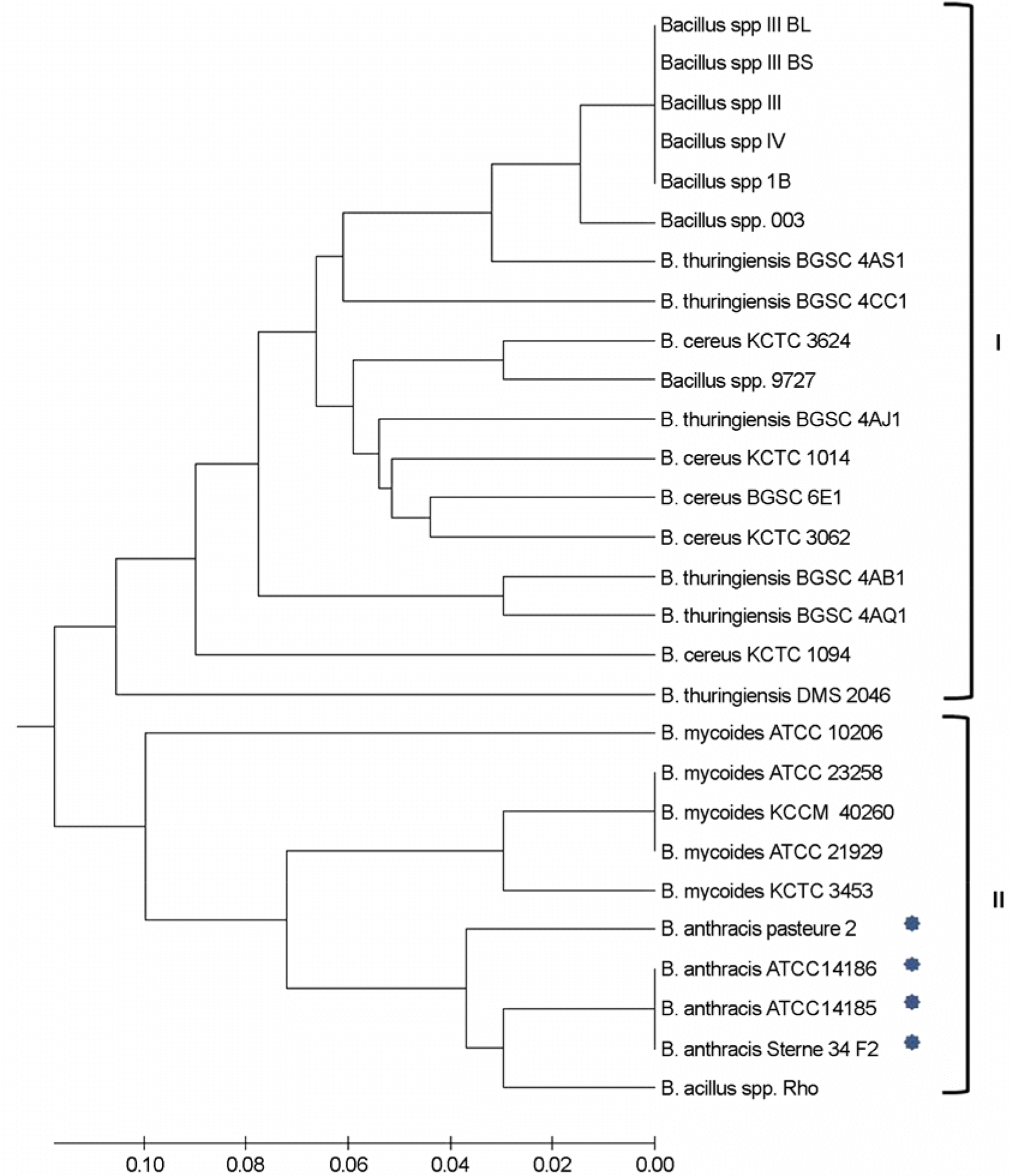
Figure 2.
MLST Dendrograms were generated by UPGMA method from genetic distances. The scale bar indicate the percentages of allelic distances between STs for MLST. The MLST dendrogram shows that B. thuringiensis BGSC 4AJ1, 4AB1, 4CC1 were the most proximate to B. anthracis.
Figure 3.
Agarose gel electrophoresis analysis of sspE gene polymerase chain reaction (PCR) on 28 strains of B. cereus group. Lane: M, 100-bp DNA size ladder; 1, B. anthracis Sterne 34-F2; 2, B. anthracis Pasteur No.2; 3, B. anthracis ATCC 14185; 4, B. anthracis ATCC 14186; 5, B. thuringiensis BGSC 4AB1; 6, B. thuringiensis BGSC 4AJ1; 7, B. thuringiensis BGSC 4AQ1; 8, B. thuringiensis BGSC 4AS1; 9, B. thuringiensis BGSC 4CC1; 10, B. thuringiensis DSM 2046; 11, B. cereus KCTC 3624; 12, B. cereus BGSC 6E1; 13, B. cereus KCTC 1014; 14, B. cereus KCTC 1094; 15, B. cereus KCTC 3062; 16, B. mycoides ATCC 10206; 17, B. mycoides ATCC 21929; 18, B. mycoides ATCC 23258; 19, B. mycoides KCCM 40260; 20, B. mycoides KCTC 3453; 21, Bacillus spp. IB; 22, Bacillus spp. 003; 23, Bacillus spp. 9727; 24, Bacillus spp. IV; 25, Bacillus spp. III; 26, Bacillus spp. III BL; 27, Bacillus spp. III BS; 28, Bacillus spp. Rho; 29, B. subtilis ATCC 6051; 30, negative water control (no template DNA).
Figure 4.
Dendrogram of 28 strains of B. cereus group constructed by UPGMA based on the bacteriological and sspE gene PCR. The clades are indicated by roman numerals. Clade III encompassed 4 strains of B. anthracis. The scale bars indicate the percentage of missmatches between biological and sspE gene PCR results.
Figure 5.
The Identification scheme of Bacillus cereus group. (First step) Gram stain, (Second step) Differenciation of Bacillus cereus group by sspE gene PCR, (Third step) Additional bacteriological tests and final identification
Table 1.
The B. cereus group strains examined in this study
Table 2.
Nucleotide sequences of the primers for the sspE gene PCR in this study
| Loci | Primers | Products (bp)a | Sequence (5′ → 3′) |
|---|---|---|---|
| sspE | sspE-F1 | 188, 71 | GAGAAAGATGAGTAAAAAACAACAA |
| (Chromosome) | sspE-R1 | CATTTGTGCTTTGAATGCTAG |
Table 3.
Cultural and biological properties of B. cereus group in this study
Table 4.
Identification Result of 28 strains by API system




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download