Abstract
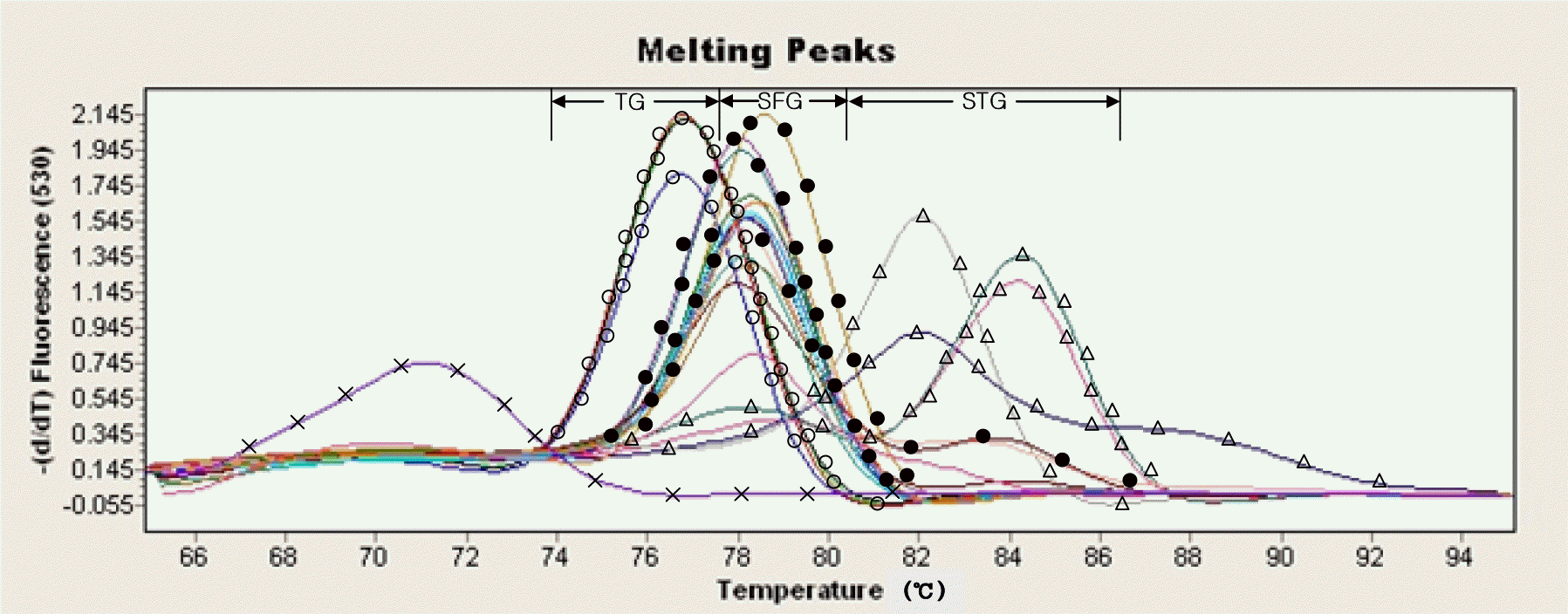
In this study, new real-time PCR method based on the groEL gene was developed and investigated. Four spotted fever group (SFG) strains, four typhus group (TG) strains, and four scrub typhus group (STG) strains were easily differentiated as a distinct entity. This PCR assay was applied to detect Rickettsia DNA from 100 ticks. Twelve Haemaphysalis longicornis ticks were found positive and identified as spotted fever group Rickettsia. This real-time PCR method could simultaneously perform the rapid identification of rickettsiae and the differential diagnosis of SFG, TG, and STG in a single reaction.
Go to : 
REFERENCES
1). Chung HY. Rickettsial Infections. Infect. 17:89–92. 1985.
2). Chung MH., Lee SH., Kim MJ., Lee JH., Kim ES., Kim MK., Park MY., Kang JS. Japanese spotted fever, South Korea. Emerg Infect Dis. 12:1122–1124. 2006.

3). Fournier PE., Roux V., Raoult D. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Int J Syst Bacteriol. 48:839–849. 1998.

4). Kim KA., Lee SH., Jang WS., Oh MD., Kim I., Choe K. Two cases of tsutsugamushi disease in the spring. Korean J Infect Dis. 31:46–49. 1999.
5). Lee JH., Park HS., Jang WJ., Koh SE., Kim JM., Shim SK., Park MY., Kim YW., Kim BJ., Kook YH., Park KH., Lee SH. Differentiation of rickettsiae by groEL gene analysis. J Clin Microbiol. 41:2952–2960. 2003.
6). Lee JH., Park HS., Jung KD., Jang WJ., Koh SE., Kang SS., Lee IY., Lee WJ., Kim BJ., Kook YH., Park KH., Lee SH. Identification of the spotted fever group rickettsiae detected from Haemaphysalis longicornis in Korea. Microbiol Immunol. 47:301–304. 2003.
7). Lee MK. Real-time polymerase chain reaction (PCR) in microbiology. Infect Chemother. 36:105–113. 2004.
8). Marston EL., Sumner JW., Regnery RL. Evaluation of intraspecies genetic variation within the 60 kDa heat-shock protein gene (groEL) of Bartonella species. Int J Syst Bacteriol. 49:1015–1023. 1999.
9). Raoult D., Dasch GA. Immunoblot cross-reactions among Rickettsia, Proteus spp. and Legionella spp. in patients with Mediterranean spotted fever. FEMS Immunol Med Microbiol. 11:13–18. 1995.
10). Raoult D., Roux V. Rickettsioses as paradigms of new or emerging infectious diseases. Clin Microbiol Rev. 10:694–719. 1997.

11). Ririe KM., Rasmussen RP., Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 245:154–160. 1997.

12). Roux V., Fournier PE., Raoult D. Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein rOmpA. J Clin Microbiol. 34:2058–2065. 1996.

13). Roux V., Raoult D. Phylogenetic analysis of the genus Rickettsia by 16S rDNA sequencing. Res Microbiol. 146:385–396. 1995.
14). Roux V., Rydkina E., Eremeeva M., Raoult D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int J Syst Bacteriol. 47:252–261. 1997.

15). Song HJ., Seong SY., Huh MS., Park SG., Jang WJ., Kee SH., Kim KH., Kim SC., Choi MS., Kim IS., Chang WH. Molecular and serologic survey of Orientia tsutsugamushi infection among field rodents in southern Cholla Province, Korea. Am J Trop Med Hyg. 58:513–518. 1998.
Go to : 
 | Figure 1.Amplification of groEL DNA from rickettsial strains using the real-time PCR. Lanes;  SFG (R japonica, R. conorii, R. sibirica, R akari), SFG (R japonica, R. conorii, R. sibirica, R akari),  : TG (R. typhi, R. typhi 87~91, R. typhi 87~100, R prowazekii), : TG (R. typhi, R. typhi 87~91, R. typhi 87~100, R prowazekii),  STG (O. tsutsugamushi Karp, O. tsutsugamushi Kato, O. tsutsugamushi Boryong, O. tsutsugamushi Gilliam), STG (O. tsutsugamushi Karp, O. tsutsugamushi Kato, O. tsutsugamushi Boryong, O. tsutsugamushi Gilliam),  : DW : DW |
 | Figure 2.Amplification of groEL DNA from rickettsial strains and tick DNA using the real-time PCR. Lanes;  : SFG (R. japonica, R. conorii, R sibirica, R akari), : SFG (R. japonica, R. conorii, R sibirica, R akari),  : TG (R. typhi, R. typhi 87~91, R. typhi 87~100, R. prowazekii), : TG (R. typhi, R. typhi 87~91, R. typhi 87~100, R. prowazekii),  : STG (O. tsutsugamushi Karp, O. tsutsugamushi Kato, O. tsutsugamushi Boryong, O. tsutsugamushi Gilliam), ————: tick (Tick no. 22, 28, 33, 36, 48, 52, 55, 61, 65, 68, 77, 92) : STG (O. tsutsugamushi Karp, O. tsutsugamushi Kato, O. tsutsugamushi Boryong, O. tsutsugamushi Gilliam), ————: tick (Tick no. 22, 28, 33, 36, 48, 52, 55, 61, 65, 68, 77, 92)  : DW : DW |
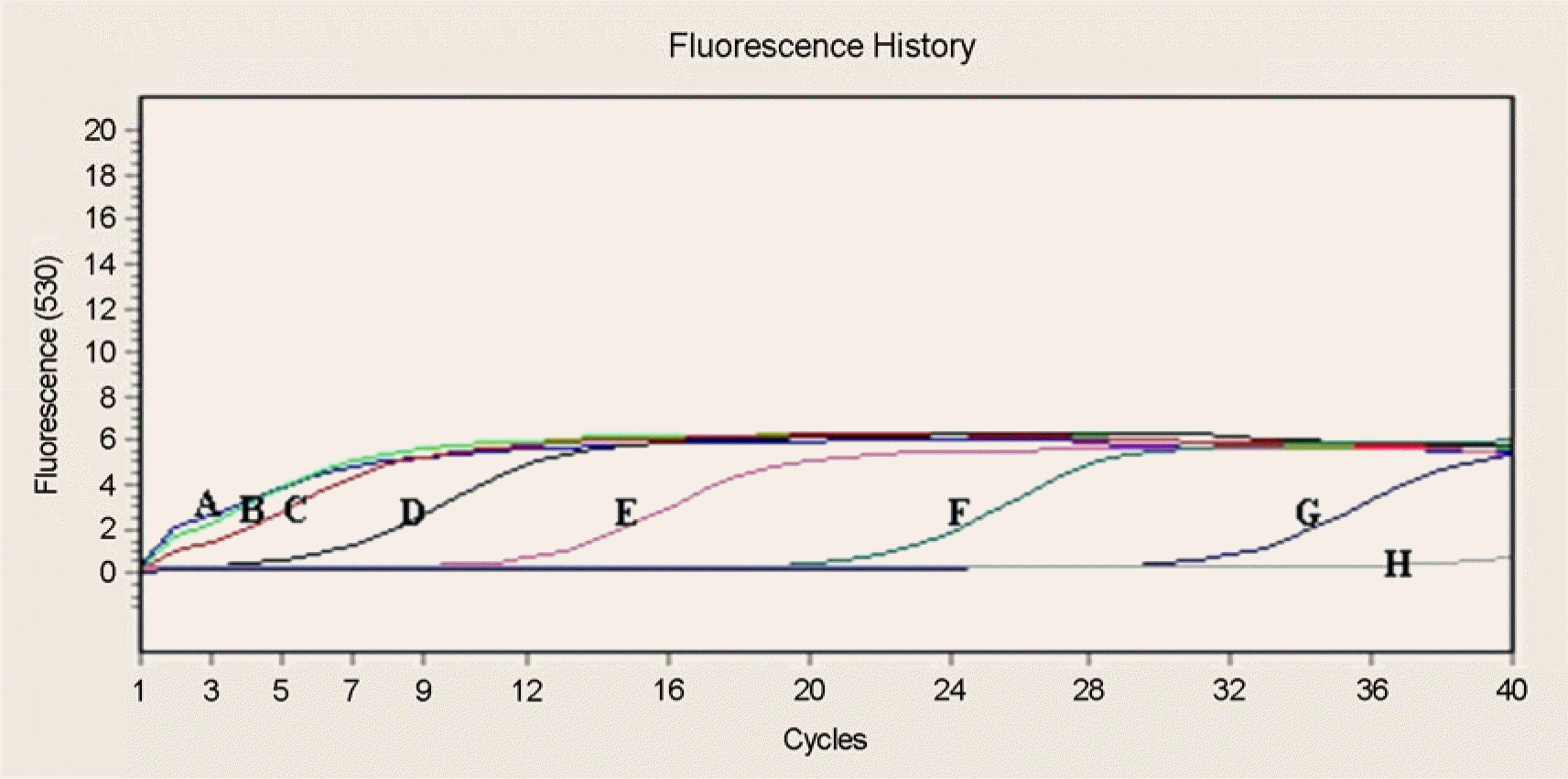 | Figure 3.The sensitivity of real-time PCR using the R. typhi plasmid. Lanes; (A) R. typhi 106, (B) R. typhi 105, (C) R. typhi 104, (D) R. typhi 103, (E) R. typhi 102, (F) R. typhi 101, (G) R. typhi 1, (H) Negative control. |
Table 1.
Rickettsial strains used in this study
| Species | Strain | Source | Geographic location | Origin (collection)a | GenBank Acession No. |
|---|---|---|---|---|---|
| groEL | |||||
| O. tsutsuga mushi | Karp | Human | New Guinea | ATCC VR-150 | M31887 |
| Kato | Human | Japan | ATCC VR-609 | AY191586 | |
| Gilliam | Human | Burma | NIH, Korea | AY191585 | |
| Boryong | Human | Korea | NIH, Korea | AY059015 | |
| R. typhi | Wilmington | Human | USA | ATCC VR-148 | AY191590 |
| R. typhi | 87~91 | Blood (human) | Korea | AY191591 | |
| R.typhi | 87~100 | Blood (human) | Korea | ||
| R. prowazekii | Breinl | Human | Poland | ATCC VR-142 | Y15783 |
| R. akari | MK | Blood (human) | USA | ATCC VR-148 | AY059013 |
| R. japonica | YH | Blood (human) | Japan | ATCC VR-1363 | AF432181 |
| R. conori | Indian Tick Typhus | Rhipiceohaluss anguineus | India | ATCC VR-597 | AY059012 |
| R. sibirica | 246 | Dermacentor nuttalli | Russia | ATCC VR-151 | AY059014 |
Table 2.
Real-time PCR condition




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download