Abstract
Mycobacterium tuberculosis-induced granulomatous lesions, particularly those undergoing central caseation, are known as hypoxic. To analyze the host genes associated with hypoxic conditions from cells infected with M. tuberculosis, we performed GeneChip analyses on mRNA from M. tuberculosis H37Rv-treated human monocytic THP-1 cells cultured in oxygen-depleted status for 18 h. The expression of 99 genes was altered, including those involved in intracellular signaling, energy production, and protein metabolism, as revealed by stringent microarray data analysis. Most notably, mRNA expression of chemokine macrophage inflammatory protein 3alpha/CC chemokine ligand 20 (CCL20) was significantly up-regulated in M. tuberculosis-infected cells under hypoxic conditions. We further analyzed the CCL20 expression in peripheral blood mononuclear cells (PBMCs) and monocyte derived macrophages (MDMs) from healthy controls and TB patients. A comparative analysis has revealed that the mRNA and protein expression of CCL20 were prominently up-regulated in PBMCs, and MDMs from TB patients, compared with healthy controls. Collectively, these data show that the gene expression of CCL20 was up-regulated in M. tuberculosis H37Rv-infected human monocytic THP-1 cells cultured in hypoxic conditions. In addition, the production of CCL20 is substantially increased in cells from TB patients than in healthy controls, suggesting an important role of CCL20 in the immunopathogenesis during TB infection.
Go to : 
References
1). Adams LB, Dinauer MC, Morgenstern DE, Krahenbuhl JL. Comparison of the roles of reactive oxygen and nitrogen intermediates in the host response to Mycobacterium tuberculosis using transgenic mice. Tuber Lung Dis. 78:237–246. 1997.
2). Bosco MC, Puppo M, Santangelo C, Anfosso L, Pfeffer U, Fardin P, Battaglia F, Varesio L. Hypoxia modifies the transcriptome of primary human monocytes: modulation of novel immune-related genes and identification of CC-chemokine ligand 20 as a new hypoxia-inducible gene. J Immunol. 177:1941–1955. 2006.

3). Burke B, Giannoudis A, Corke KP, Gill D, Wells M, Ziegler-Heitbrock L, Lewis CE. Hypoxia-induced gene expression in human macrophages: implications for ischemic tissues and hypoxia-regulated gene therapy. Am J Pathol. 163:1233–1243. 2003.
4). Cazin M, Paluszezak D, Bianchi A, Cazin JC, Aerts C, Voisin C. Effects of anaerobiosis upon morphology and energy metabolism of alveolar macrophages cultured in gas phase. Eur Respir J. 3:1015–1022. 1990.
5). Cook DN, Prosser DM, Forster R, Zhang J, Kuklin NA, Abbondanzo SJ, Niu XD, Chen SC, Manfra DJ, Wiekowski MT, Sullivan LM, Smith SR, Greenberg HB, Narula SK, Lipp M, Lira SA. CCR6 mediates dendritic cell localization, lymphocyte homeostasis, and immune responses in mucosal tissue. Immunity. 12:495–503. 2000.

6). Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, Orme IM. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J Exp Med. 178:2243–2247. 1993.

7). Cooper AM, Segal BH, Frank AA, Holland SM, Orme IM. Transient loss of resistance to pulmonary tuberculosis in p47(phox–/–) mice. Infect Immun. 68:1231–1234. 2000.
8). Demissie A, Abebe M, Aseffa A, Rook G, Fletcher H, Zumla A, Weldingh K, Brock I, Andersen P, Doherty TM. VACSEL Study Group: Healthy individuals that control a latent infection with Mycobacterium tuberculosis express high levels of Th1 cytokines and the IL-4 antagonist IL-4 delta2. J Immunol. 172:6938–6943. 2004.
9). Dieu MC, Vanbervliet B, Vicari A, Bridon JM, Oldham E, Ait-Yahia S, Briere F, Zlotnik A, Lebecque S, Caux C. Selective recruitment of immature and mature dendritic cells by distinct chemokines expressed in different anatomic sites. J Exp Med. 188:373–386. 1998.

10). Dye C, Scheele S, Dolin P, Pathania V, Raviglione MC. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA. 282:677–686. 1999.
11). Fenton MJ, Vermeulen MW. Immunopathology of tuberculosis: roles of macrophages and monocytes. Infect Immun. 64:683–690. 1996.

14). Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med. 178:2249–2254. 1993.
15). Greaves DR, Wang W, Dairaghi DJ, Dieu MC, Saint-Vis B, Franz-Bacon K, Rossi D, Caux C, McClanahan T, Gordon S, Zlotnik A, Schall TJ. CCR6, a CC chemokine receptor that interacts with macrophage inflammatory protein 3alpha and is highly expressed in human dendritic cells. J Exp Med. 186:837–844. 1997.
16). Homey B, Dieu-Nosjean MC, Wiesenborn A, Massacrier C, Pin JJ, Oldham E, Catron D, Buchanan ME, Muller A, deWaal Malefyt R, Deng G, Orozco R, Ruzicka T, Lehmann P, Lebecque S, Caux C, Zlotnik A. Up-regulation of macrophage inflammatory protein-3 alpha/CCL20 and CC chemokine receptor 6 in psoriasis. J Immunol. 164:6621–6632. 2000.
17). Honer zu Bentrup K, Russell DG. Mycobacterial persistence: adaptation to a changing environment. Trends Microbiol. 9:597–605. 2001.
18). Kao CY, Huang F, Chen Y, Thai P, Wachi S, Kim C, Tam L, Wu R. Up-regulation of CC chemokine ligand 20 expression in human airway epithelium by IL-17 through a JAK-independent but MEK/NF-kappaB-dependent signaling pathway. J Immunol. 175:6676–6685. 2005.
19). Kawaguchi T, Veech RL, Uyeda K. Regulation of energy metabolism in macrophages during hypoxia. Roles of fructose 2,6-bisphosphate and ribose 1,5-bisphosphate. J Biol Chem. 276:28554–28561. 2001.
20). Lee JH, Kang HJ, Woo JS, Chae SW, Lee SH, Hwang SJ, Lee HM. Up-regulation of chemokine ligand 20 in chronic rhinosinusitis. Arch Otolaryngol Head Neck Surg. 132:537–541. 2006.

21). Lee JS, Son JW, Jung SB, Kwon YM, Yang CS, Oh JH, Song CH, Kim HJ, Park JK, Paik TH, Jo EK. Ex vivo responses for interferon-gamma and proinflammatory cytokine secretion to low-molecular-weight antigen MTB12 of Mycobacterium tuberculosis during human tuberculosis. Scand J Immunol. 64:145–154. 2006.
22). Lee JS, Song CH, Lim JH, Kim HJ, Park JK, Paik TH, Kim CH, Kong SJ, Shon MH, Jung SS, Jo EK. The production of tumour necrosis factor-alpha is decreased in peripheral blood mononuclear cells from multidrug-resistant tuberculosis patients following stimulation with the 30-kDa antigen of Mycobacterium tuberculosis. Clin Exp Immunol. 132:443–449. 2003.
23). Parrish NM, Dick JD, Bishai WR. Mechanisms of latency in Mycobacterium tuberculosis. Trends Microbiol. 6:107–112. 1998.
24). Rubie C, Frick VO, Wagner M, Rau B, Weber C, Kruse B, Kempf K, Tilton B, Konig J, Schilling M. Enhanced expression and clinical significance of CC-chemokine MIP-3 alpha in hepatocellular carcinoma. Scand J Immunol. 63:468–477. 2006.
25). Schutyser E, Struyf S, Van Damme J. The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev. 14:409–426. 2003.

26). Sugita S, Kohno T, Yamamoto K, Imaizumi Y, Nakajima H, Ishimaru T, Matsuyama T. Induction of macrophage-inflammatory protein-3 alpha gene expression by TNF-α dependent NF-kappa B activation. J Immunol. 168:5621–5628. 2002.
27). Song CH, Lee JS, Lee SH, Lim K, Kim HJ, Park JK, Paik TH, Jo EK. Role of mitogen-activated protein kinase pathways in the production of tumor necrosis factor-alpha, interleukin-10, and monocyte chemotactic protein-1 by Mycobacterium tuberculosis H37Rv-infected human monocytes. J Clin Immunol. 23:194–201. 2003.
28). Ulrichs T, Kaufmann SH. New insights into the function of granulomas in human tuberculosis. J Pathol. 208:261–269. 2006.

29). Volpe E, Cappelli G, Grassi M, Martino A, Serafino A, Colizzi V, Sanarico N, Mariani F. Gene expression profiling of human macrophages at late time of infection with Mycobacterium tuberculosis. Immunology. 118:449–460. 2006.
30). Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 64:2062–2069. 1996.
31). Werngren J, Hoffner SE. Drug-susceptible Mycobacterium tuberculosis Beijing genotype does not develop mutation-conferred resistance to rifampin at an elevated rate. J Clin Microbiol. 41:1520–1524. 2003.
32). Yang D, Chen Q, Hoover DM, Staley P, Tucker KD, Lubkowski J, Oppenheim JJ. Many chemokines including CCL20/ MIP-3 alpha display antimicrobial activity. J Leukoc Biol. 74:448–455. 2003.
Go to : 
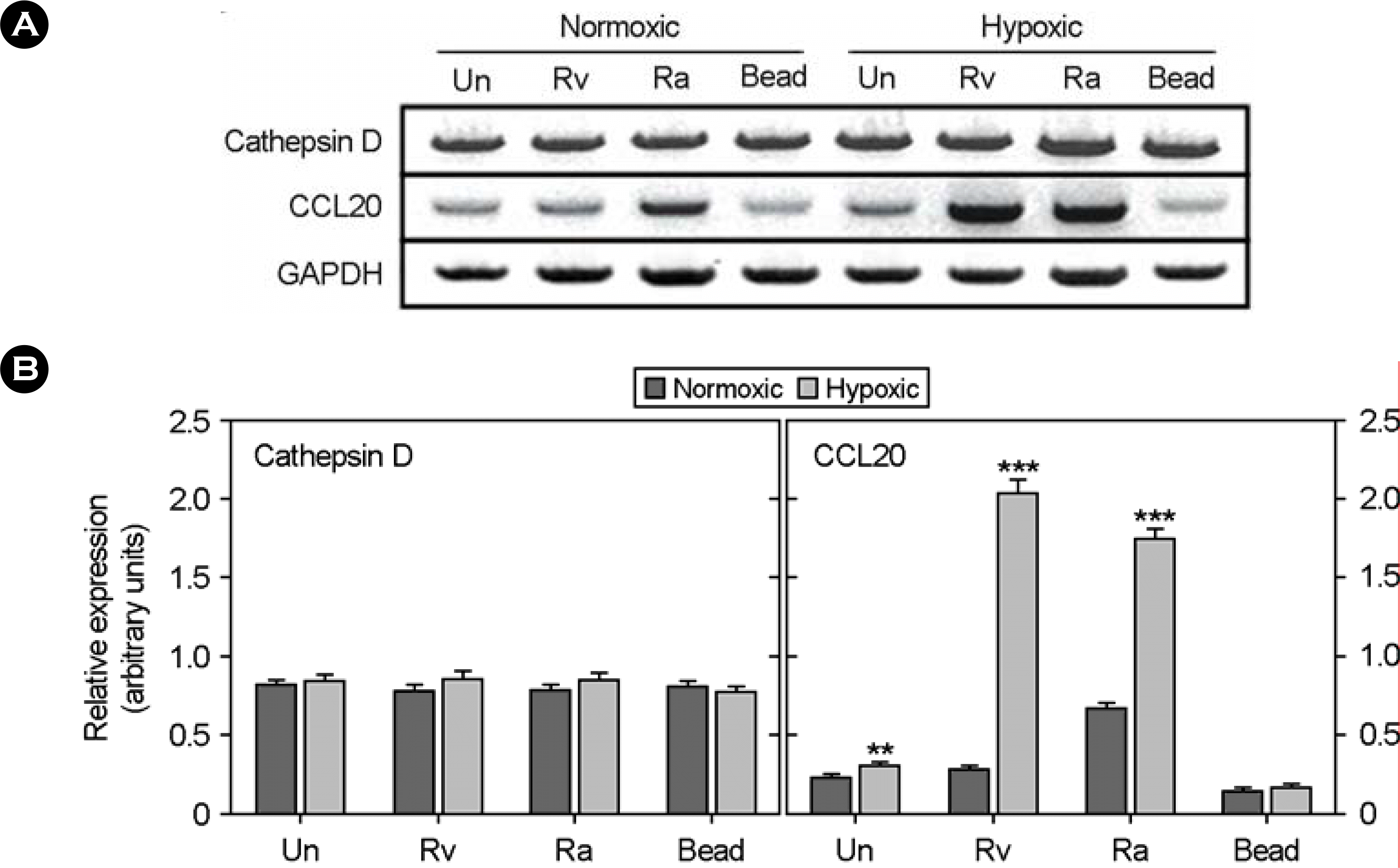 | Figure 1.Comparative analysis of human cathepsin D and CCL20 mRNA expression in M. tuberculosis-infected THP-1 cells under normoxic and hypoxic conditions. (A) RT-PCR analysis was used to determine the differential expression of cathepsin D and CCL20 genes in THP-1 cells cultured under normoxic and hypoxic conditions. normoxic, aerobically incubated (20% O2); Hypoxic, incubated in hypoxic conditions (2% O2); Un, unstimluated. Rv, M. tuberculosis H37Rv-infected; Ra, M. tuberculosis H37Ra-infected. (B) Values are ratios of band density to band density of GAPDH mRNA at each condition and are mean ± SEM. ∗∗, p<0.01; ∗∗∗, p<0.001 |
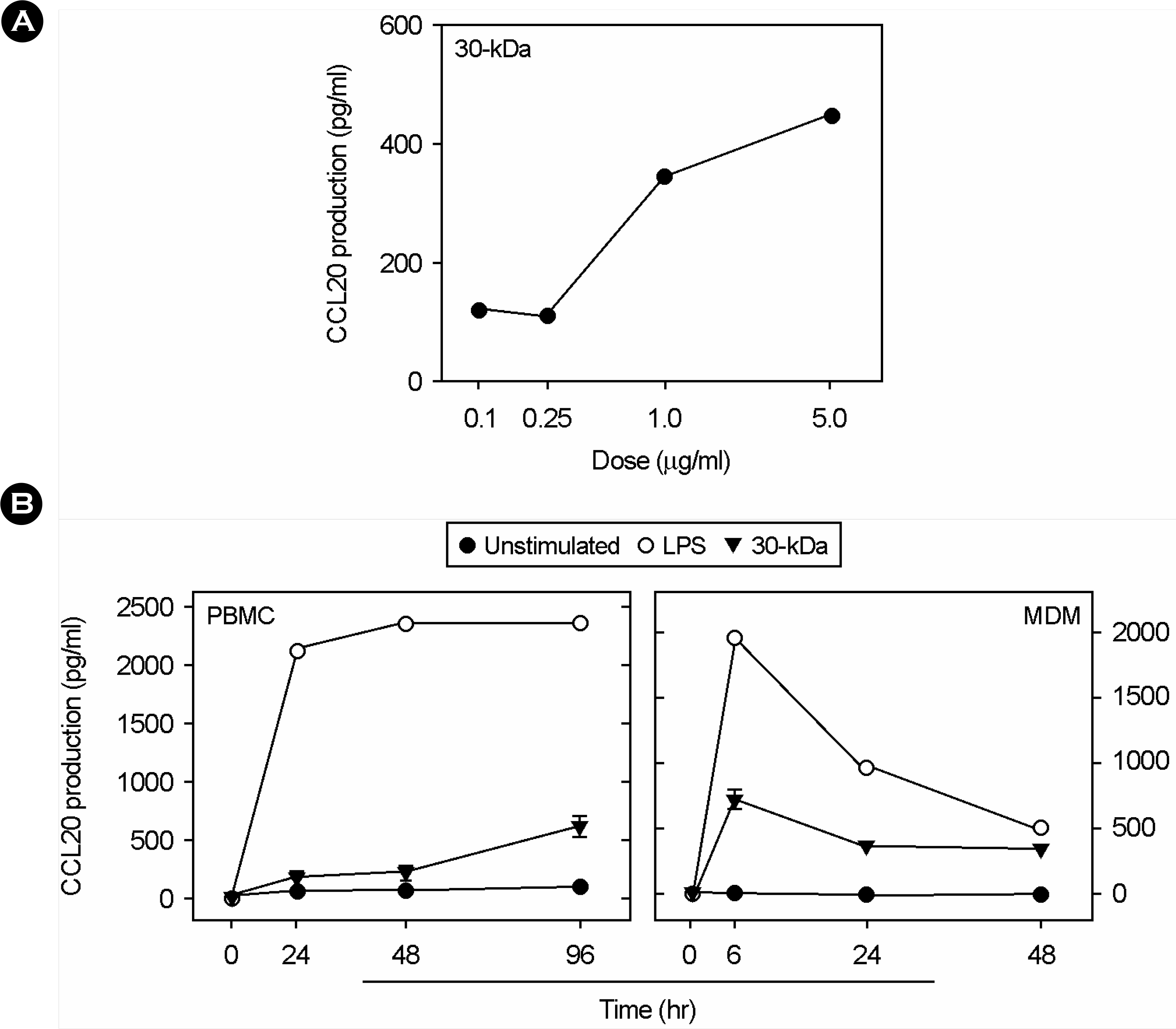 | Figure 2.Kinetics of CCL20 induction in response to 30-kDa Ag. PBMCs and MDMs were incubated with different doses of 30-kDa Ag (A) and different time point (B). Supernatants were harvested after the times indicated and CCL20 levels were measured by ELISA. |
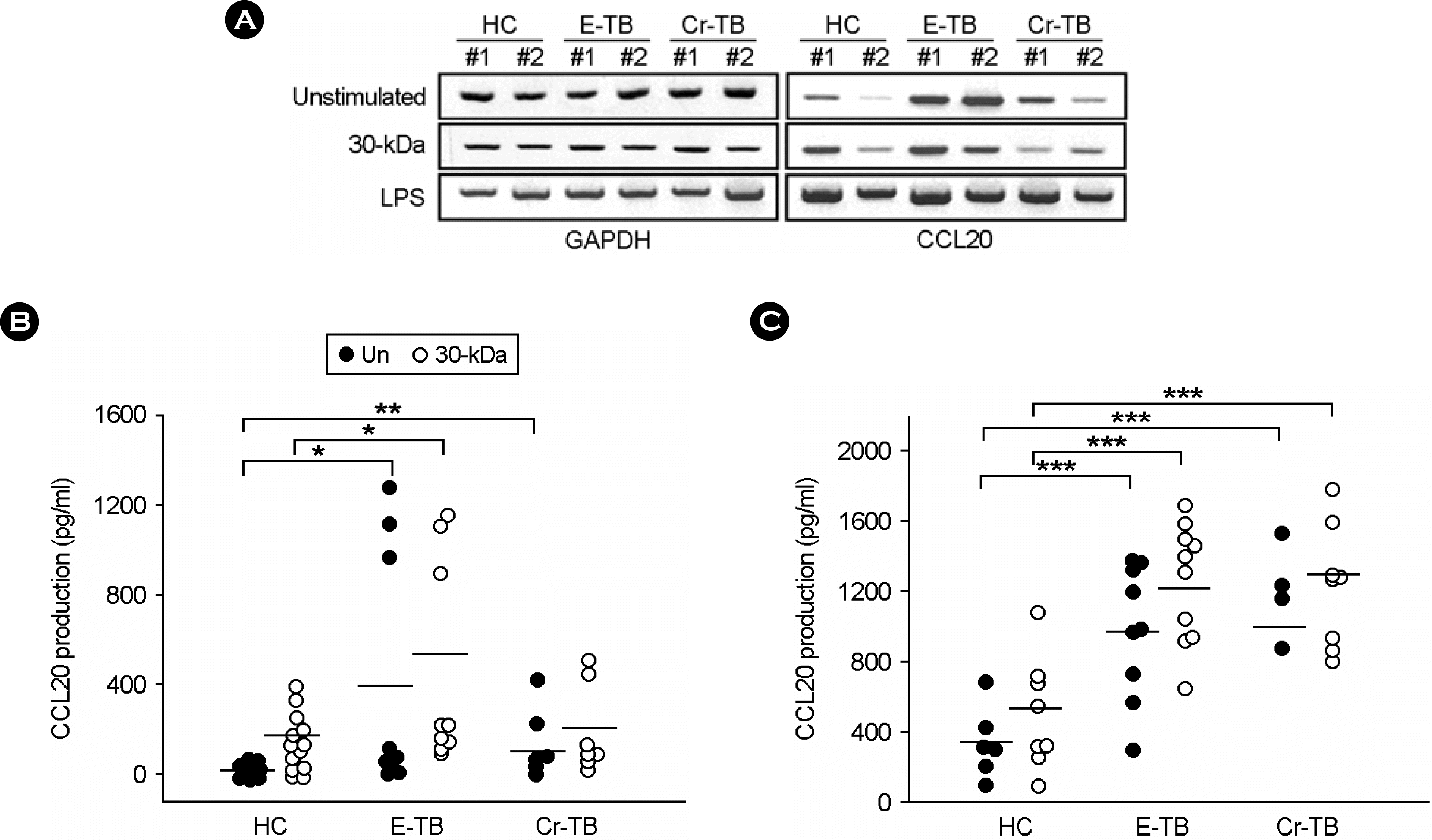 | Figure 3.CCL20 expressions in PBMCs and MDMs from healthy controls (HC) and TB patients (E-TB and Cr-TB) in response to 30-kDa Ag. (A) The 30-kDa Ag was added to PBMCs at 1.0 μg/ml for 6 hr. RT-PCR analysis was done to determine the comparative expression of CCL20 mRNA from PBMCs from HCs and TB patients. PBMCs (B) and MDMs (C) from HCs and TB patients were stimulated by 30-kDa Ag. After 18-hr stimulation, the supernatants were harvested and CCL20 protein levels were measured by ELISA. Closed circle, unstimulated; open circle, stimulated with the 30-kDa Ag. E-TB, early diagnosed pulmonary tuberculosis patients; Cr-TB, chronic tuberculosis patients. ∗, p<0.05; ∗∗, p<0.01; ∗∗∗, p<0.001 |
Table 1.
Down-regulated genes in THP-1 cells infected with M. tuberculosis H37Rv under hypoxic conditions
| Genbank accession no. | Symbol | Name of gene | Fold changea |
|---|---|---|---|
| 1. Binding | |||
| R43015 | SF3A3 | Splicing factor 3a, subunit 3, 60kDa | 2.14 |
| AA047478 | CORO1A | Coronin, actin binding protein, 1A | 2.57 |
| AA281733 | EIF1AX | Eukaryotic translation initiation factor 1A, X-linked | 2.30 |
| AA460286 | GNG10 | Guanine nucleotide binding protein (G protein), gamma 10 | 2.10 |
| AA133577 | SNRPG | Small nuclear ribonucleoprotein polypeptide G | 2.57 |
| W72693 | HNRPAB | Heterogeneous nuclear ribonucleoprotein A/B | 2.92 |
| AA454585 | SFRS2 | Splicing factor, arginine/serine-rich 2 | 2.31 |
| AA453749 | HDGF | Hepatoma-derived growth factor | 2.82 |
| AA464731 | S100A11 | S100 calcium binding protein A11 (calgizzarin) | 2.02 |
| AA598526 | HIF1A | Hypoxia-inducible factor 1, alpha subunit | 3.41 |
| AW075424 | SFRS3 | Splicing factor, arginine/serine-rich 3 | 2.60 |
| AA086471 | S100A8 | S100 calcium binding protein A8 (calgranulin A) | 5.78 |
| AW087572 | CALM2 | Calmodulin 2 (phosphorylase kinase, delta) | 2.65 |
| 2. Catalytic activity | |||
| T72398 | TDO2 | Tryptophan 2,3-dioxygenase | 3.58 |
| AW028938 | PPM1B | Protein phosphatase 1B (formerly 2C) | 4.23 |
| H65395 | PSME2 | Proteasome (prosome, macropain) activator subunit 2 (PA28 beta) | 2.38 |
| AA863149 | PSMA7 | Proteasome (prosome, macropain) subunit, alpha type, 7 | 2.01 |
| T71606 | HMOX1 | Heme oxygenase (decycling) 1 | 2.17 |
| AI375353 | SGK | Serum/glucocorticoid regulated kinase | 2.21 |
| N20475 | CTSD | Cathepsin D (lysosomal aspartyl protease) | 7.23 |
| AI361530 | ACSL1 | Acyl-CoA synthetase long-chain family member 1 | 2.55 |
| AA708298 | ATP5B | ATP synthase, H+ transporting, mitochondrial F1 complex | 2.54 |
| AA160913 | PDE9A | Phosphodiesterase 9A | 2.39 |
| AI828190 | PLAUR | Plasminogen activator, urokinase receptor | 2.15 |
| AW087572 | CALM2 | Calmodulin 2 (phosphorylase kinase, delta) | 2.65 |
| AW078798 | UBB | Ubiquitin B | 2.38 |
| 3. Chaperone activity | |||
| AA448396 | HSPE1 | Heat shock 10 kDa protein 1 (chaperonin 10) | 2.40 |
| AW075457 | CCT3 | Chaperonin containing TCP1, subunit 3 (gamma) | 2.70 |
| AW004895 | HSPD1 | Heat shock 60 kDa protein 1 (chaperonin) | 3.40 |
| 4. Defense immunity protein activity | |||
| AA865464 | LY6E | Lymphocyte antigen 6 complex, locus E | 2.35 |
| H29485 | SSB | Sjogren syndrome antigen B (autoantigen La) | 2.13 |
| AI362062 | NOVA1 | Neuro-oncological ventral antigen 1 | 2.06 |
| AA775355 | XRCC5 | X-ray repair complementing defective repair in CHO cells 5 | 2.26 |
| AW075443 | G22P1 | Thyroid autoantigen 70 kDa (Ku antigen) | 2.25 |
| 5. Signal transducer activity | |||
| AW058480 | LBR | Lamin B receptor | 2.47 |
| AA133212 | NCOA4 | Nuclear receptor coactivator 4 | 2.21 |
| AI828190 | PLAUR | Plasminogen activator, urokinase receptor | 2.15 |
| 6. Transporter activity | |||
| H85355 | ATP2A | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 2.26 |
| AI815076 | 2SLC7A7 | Solute carrier family 7 (cationic amino acid transporter) | 2.17 |
| R52654 | CYCS | Cytochrome c, somatic | 2.99 |
| AA598759 | PGD | Phosphogluconate dehydrogenase | 2.27 |
| AI969670 | LDHB | Lactate dehydrogenase B | 2.68 |
| AA708298 | ATP5B | ATP synthase, H+ transporting, mitochondrial F1 complex | 2.54 |
| AA865265 | CYCS | Cytochrome c, somatic | 2.47 |
| AA463492 | CYBB | Cytochrome b-245, beta polypeptide | 2.07 |
| 7. Others | |||
| AA608568 | CCNA2 | Cyclin A2 | 2.65 |
| H99170 | CALR | Calreticulin | 2.70 |
| AA699697 | TNF | Tumor necrosis factor (TNF superfamily, member 2) | 2.02 |
| AA644088 | CTSC | Cathepsin C | 2.00 |
| AA406020 | G1P2 | Interferon, alpha-inducible protein (clone IFI-15K) | 2.13 |
| AA600177 | CALR | Calreticulin | 2.08 |
| AA489087 | KPNA2 | Karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | 3.82 |
| W73874 | CTSL | Cathepsin L | 2.70 |
| AW087897 | OK/SW-cl.56 | Beta 5-tubulin | 2.22 |
| AW071125 | PRDX1 | Peroxiredoxin 1 | 3.39 |
| W73144 | LCP1 | Lymphocyte cytosolic protein 1 (L-plastin) | 2.42 |
| N90109 | NCL | Nucleolin | 4.84 |
Table 2.
Up-regulated genes in THP-1 cells infected with M. tuberculosis H37Rv under hypoxic conditions
| Genbank accession no. | Symbol | Name of gene | Fold changea |
|---|---|---|---|
| 1. Binding | |||
| AA936135 | Gemin7 | Gem (nuclear organelle) associated protein 7 | 2.40 |
| AA278764 | ZNF406 | Zinc finger protein 406 | 3.24 |
| AI800882 | NRL | Neural retina leucine zipper | 2.41 |
| AA457153 | ZNF282 | Zinc finger protein 282 | 2.09 |
| 2. Catalytic activity | |||
| AA644211 | PTGS2 | Prostaglandin-endoperoxide synthase 2 | 6.93 |
| AA459292 | CKS1B | CDC28 protein kinase regulatory subunit 1B | 2.74 |
| AI961583 | MAPK10 | Mitogen-activated protein kinase 10 | 2.15 |
| AA496013 | NEK4 | NIMA (never in mitosis gene a)-related kinase 4 | 2.05 |
| R61229 | GATM | Glycine amidinotransferas | 3.57 |
| AA427725 | CPZ | Carboxypeptidase Z | 2.10 |
| AA481562 | DARS | Aspartyl-tRNA synthetase | 2.07 |
| AA457671 | P4HA1 | Procollagen-proline, 2-oxoglutarate 4-dioxygenase | 4.35 |
| AA630620 | PIK3R4 | Phosphoinositide-3-kinase, regulatory subunit 4, p150 | 2.59 |
| AA457700 | SCD | Stearoyl-CoA desaturase (delta-9-desaturase) | 3.34 |
| AA279533 | HMGCS1 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 | 3.61 |
| AA461478 | RNGTT | RNA guanylyltransferase and 5′-phosphatase | 2.04 |
| AA866043 | CBS | Cystathionine-beta-synthase | 2.01 |
| N63567 | PDK3 | Pyruvate dehydrogenase kinase, isoenzyme 3 | 2.00 |
| AI335255 | NEK6 | NIMA (never in mitosis gene a)-related kinase 6 | 2.05 |
| 3. Cell adhesion molecule | |||
| AA922832 | ICAM3 | Intercellular adhesion molecule 3 | 2.13 |
| 4. Signal transducer activity | |||
| AA453774 | RGS16 | Regulator of G-protein signalling 16 | 4.95 |
| T62636 | CXCR4 | Chemokine (C-X-C motif) receptor 4 | 3.06 |
| AA017544 | RGS1 | Regulator of G-protein signalling 1 | 5.08 |
| AI380755 | PVR | Poliovirus receptor | 2.08 |
| 5. Transcription regulator activity | |||
| AI418194 | SOX21; SOX25LD | Homo sapiens cDNA clone IMAGE: 2105121 3′, mRNA sequence | 2.25 |
| 6. Transporter activity | |||
| AA453467 | LDHC | Lactate dehydrogenase C | 2.22 |
| AA157955 | SC4MOL | Sterol-C4-methyl oxidase-like | 2.40 |
| AI160757 | KCNJ10C | Potassium inwardly-rectifying channel, subfamily J, member 10 | 2.73 |
| 7. Others | |||
| AI285199 | CL20 | Chemokine (C-C motif) ligand 20 | 5.54 |
| AA857944 | CSPG2 | Chondroitin sulfate proteoglycan 2 (versican) | 7.76 |
| AA450123 | ENO2 | Enolase 2 (gamma, neuronal) | 4.90 |
| AI951114 | ALDOC | Aldolase C, fructose-bisphosphate | 4.43 |
| AI656802 | OLFM1 | Olfactomedin 1 | 2.12 |
| AA872001 | ANXA6 | Annexin A6 | 2.21 |
| AA620859 | SSPN | Sarcospan (Kras oncogene-associated gene) | 2.01 |
| AI362949 | SYN2 | Synapsin II | 2.36 |
| AI056417 | MSLN | Mesothelin | 2.12 |
| AI015641 | SDC1 | Syndecan 1 | 2.27 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download