Abstract
Attaching and effacing Escherichia coli (AEEC) cause enteric infections in humans and animals. Attaching indicates the intimate attachment of bacteria to the enterocyte, and effacing relates to the localized effacement of brush border microvilli. Enteropathogenic (EPEC) and enterohemorrhagic Escherichia coli (EHEC) infections are characterized by the formation of attaching and effacing (AE) lesion on the intestinal epithelial cells. Therefore, they are often grouped together as AEEC. Development of multiplex PCR allowed us to type five of the most important genes implicated in the formation of the AE lesion. A total of 60 AEEC strains isolated from diarrheal patients were investigated by multiplex PCR for the presence of the insertion site of locus of enterocyte effacement (LEE) and LEE-related (eae, tir, espA, espB, and espD) genes. Associating the results of LEE genes typing in the AEEC strains, three different pathotypes are determined: eaeγ-tirγ-espAγ-espBγ-espDγ (O157:H7), eaeβ-tirβ-espAβ-espBβ-espDβ (O26:H11), and eaeα-tirα-espAα-espBα-espDα (O55:H6). These results indicate that AEEC are a heterogenous groups of organisms.
References
1). 김도균, 이상래, 김정우. 장출혈성 대장균 O157:H7 유 래 재조합 intimin의 발현과 그의 면역반응 효과. 동물 자원지. 46:495–502. 2004.
2). 김선희, 송현제, 정재근, 하동룡, 류필열, 이종빈. 광주 지역에서 분리된 장병원성 대장균의 표현형 및 유전자형 특성에 관한 연구. J Bac Virol. 36:167–174. 2006.
3). 김영부. 설사환자에 분리한 장관출혈성 대장균 O157: H7의 병원인자와 Arbitrarily-primed polymerase chain reaction의 형별. J Bac Virol. 31:123–131. 2001.
4). 손원근, Gannon VP. Escherichia coli O157:H7 intimin의 expression 및 C-terminal 부위의 특징. Kor J Vet Publ Hlth. 25:133–140. 2001.
5). Adu-Bobie J, Frankel G, Bain C, Goncalves AG, Trabulsi LR, Douce G, Knutton S, Dougan G. Detection of intimins alpha, beta, gamma, and delta, four intimin derivatives expressed by attaching and effacing microbial pathogens. J Clin Microbiol. 36:662–668. 1998.
6). An H, Fairbrother JM, Dubreuil D, Harel J. Cloning and characterization of the eae gene from a dog attaching and effacing Escherichia coli strain 4221. FEMS Microbiol Lett. 148:239–245. 1997.
7). Boerlin P, Chen S, Colbourne JK, Johnson R, De Grandis S, Gyles C. Evolution of enterohemorrhagic Escherichia coli hemolysin plasmids and the locus for enterocyte effacement in shiga toxin-producing E. coli. Infect Immun. 66:2553–2561. 1998.
8). Cebula TA, Payne WL, Feng P. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J Clin Microbiol. 33:248–250. 1995.
9). China B, Devrin AC, Jacquemin E, Pirson V, Mainil J. Heterogeneity of the eae genes in attaching/effacing Escherichia coli from cattle: comparison with human strains. Res Microbiol. 150:323–332. 1999.
10). China B, Goffaux F, Pirson V, Mainil J. Comparison of eae, tir, espA and espB genes of bovine and human attaching and effacing Escherichia coli by multiplex polymerase chain reaction. FEMS Microbiol Lett. 178:177–182. 1999.
11). Donnenberg MS, Lai LC, Taylor KA. The locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli encodes secretion functions and remnants of transposons at its extreme right end. Gene. 184:107–114. 1997.
12). Donnenberg MS, Tacket CO, James SP, Losonsky G, Nataro JP, Wasserman SS, Kaper JB, Levine MM. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J Clin Invest. 92:1412–1417. 1993.
13). Donnenberg MS, Tacket CO, Losonsky G, Frankel G, Nataro JP, Dougan G, Levine MM. Effect of prior experimental human enteropathogenic Escherichia coli infection on illness following homologous and heterologous rechallenge. Infect Immun. 66:52–58. 1998.
14). Donnenberg MS, Tzipori S, McKee ML, O'Brien AD, Alroy J, Kaper JB. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J Clin Invest. 92:1418–1424. 1993.
15). Elliott SJ, Wainwright LA, McDaniel TK, Jarvis KG, Deng YK, Lai LC, McNamara BP, Donnenberg MS, Kaper JB. The complete sequence of the locus of enteroycte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol. 28:1–4. 1998.
16). Finlay BB, Rosenshine I, Donnenberg MS, Kaper JB. Cytoskeletal composition of attaching and effacing lesions associated with enteropathogenic Escherichia coli adherence to HeLa cells. Infect Immun. 60:2541–2543. 1992.
17). Gannon VP, Rashed M, King RK, Thomas EJ. Detection and characterization of the eae gene of Shiga-like toxin-producing Escherichia coli using polymerase chain reaction. J Clin Microbiol. 31:1268–1274. 1993.
18). Giron JA, Donnenberg MS, Martin WC, Jarvis KG, Kaper JP. Distribution of the bundle-forming pilus structural gene (bfpA) among enteropathogenic Escherichia coli. J Infect Dis. 168:1037–1041. 1993.
19). Goffaux F, China B, Janssen L, Mainil J. Genotypic characterization of enteropathogenic Escherichia coli (EPEC) isolated in Belgium from dogs and cats. Res Microbiol. 151:865–871. 2000.
20). Goffaux F, Mainil J, Pirson V, Charlier G, Pohl P, Jacquemin E, China B. Bovine attaching and effacing Escherichia coli possess a pathogenesis island related to the LEE of the human enteropathogenic E. coli strain E2348/69. FEMS Microbiol Lett. 154:415–421. 1997.
21). Gyles CL. Escherichia coli cytotoxins and enterotoxins. Can J Microbiol. 38:734–746. 1992.
22). Honda T. Factors influencing the development of hemolytic uremic syndrome caused by enterohemorrhagic Escherichia coli infection: from a questionnaire survey to in vitro experiment. Pediatr Int. 41:209–212. 1999.
23). Imberechts H, De Greve H, Lintermans P. The pathogenesis of edema disease in swine. Vet Microbiol. 31:221–233. 1992.
24). Jarvis KG, Giron JA, Jerse AE, McDaniel TK, Donnenberg MS, Kaper JB. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci USA. 92:7996–8000. 1995.
25). Jarvis KG, Kaper JB. Secretion of extracellular proteins by enterohemorrhagic Escherichia coli via a putative type III secretion system. Infect Immun. 64:4826–4829. 1996.
26). Jerse AE, Yu J, Tall BD, Kaper JB. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA. 87:7839–7843. 1990.
27). Kaper JB, Elliott S, Sperandio V, Perna NT, Mayhew GF, Blattner FR. Attaching and effacing intestinal histopathology and the locus of enterocyte effacement. Kaper JB, O'Brien AD, editors. (Eds.):. Escherichia coli O157:H7 and Other Shiga Toxin-Producing E. coli Strains. American Society for Microbiology;Washington, DC: p. 163–182. 1998.
28). Karaolis DKR, McDaniel TK, Kaper JB, Boedeker EC. Cloning of the RDEC-1 locus of enterocyte effacement (LEE) and functional analysis of the phenotype on Hep-2 cells. Adv Exp Med Biol. 412:241–245. 1997.

29). Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. Enteropathogenic (EPEC) E. coli transfers its receptor for intimate adherence into mammalian cells. Cell. 91:511–520. 1997.
30). Kim YB, Okuda J, Matsumoto C, Morigaki T, Asai N, Watanabe H, Nishibuchi M. Isolation of an Escherichia coli O157:H7 strain producing Shiga toxin 1 but not Shiga toxin 2 from a patient with hemolytic uremic syndrome in Korea. FEMS Microbiol Lett. 166:43–48. 1998.
31). Knutton S. Attaching and effacing E. coli. Gyles CL, editor. (Ed.),. Escherichia coli in Domestic Animals and Humans. CAB International;Wallingford: p. 567–591. 1994.
32). Knutton S. Cellular responses to enteropathogenic Escherichia coli infection. Biosci Rep. 15:469–479. 1995.
33). Kresse AU, Beltrametti F, Muller A, Ebel F, Guzman CA. Characterization of SepL of enterohemorrhagic Escherichia coli. J Bacteriol. 182:6490–6498. 2000.
34). Lai LC, Wainwright LA, Stone KD, Donnenberg MS. A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect Immun. 65:2211–2217. 1997.
35). McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 92:1664–1668. 1995.

36). Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol Microbiol. 33:296–306. 1999.
37). Nataro JP, Kaper JB. Diarrhoeagic Escherichia coli. Clin Microbiol Rev. 11:142–201. 1998.
38). Osek J. Development of a multiplex PCR approach for the identification of Shiga toxin-producing Escherichia coli strains and their major virulence factor genes. J Appl Microbiol. 95:1217–1225. 2003.
39). Paton AW, Mannig PA, Woodrow MC, Paton JC. Translocated intimin receptor (Tir) of Shiga-toxigenic Escherichia coli isolates belonging to serogroups, O26, O111, and O157 react with sera from patients with hemolytic-Uremic syndrome and exhibit marked sequence heterogeneity. Infect Immun. 66:5580–5586. 1998.
40). Perna N, Mayhew GF, Posfai G, Elliott S, Donnenberg MS, Kaper JB, Blattner FR. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 66:3810–3817. 1998.
41). Schmidt H, Scheff J, Morabito S, Caprioli A, Wieler LH, Karch H. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl Environ Microbiol. 66:1205–1208. 2000.
42). Wang G, Clark CG, Rodgers FG. Detection of Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J Clin Microbiol. 40:3613–3619. 2002.
43). Zhu C, Agin TS, Elliott SJ, Johnson LA, Thate TE, Kaper JB, Boedeker EC. Complete nucleotide sequence and analysis of the locus of enterocyte effacement from rabbit diarrheagenic Escherichia coli RDEC-1. Infect Immun. 69:2107–2115. 2001.
44). Zweifel C, Blanco JE, Blanco M, Blanco J, Stephan R. Serotypes and virulence genes of ovine non-O157 Shiga toxin-producing Escherichia coli in Switzerland. Int J Food Microbiol. 95:19–27. 2004.
Figure 1.
stx genes obtained from AEEC strains by multiplex PCR A∼C. A: lane M, 100 bp plus ladder (molecular weight markers); lane 1, control strains EDL933 (stx1+, stx2+); lane 2, EHEC O1157:H– strain (stx1–, stx2+); lane 3, EHEC O111:H– strain (stx1+, stx2–); lane 4, EPEC O55:H– strain (stx1–, stx2–). B: lane N, 100 bp ladder (molecular weight markers); lane 1, E. coli EDL933 (stx2e–, stx2f +); lane 2, EHEC O1157:H–strain (stx2e–, stx2f+).
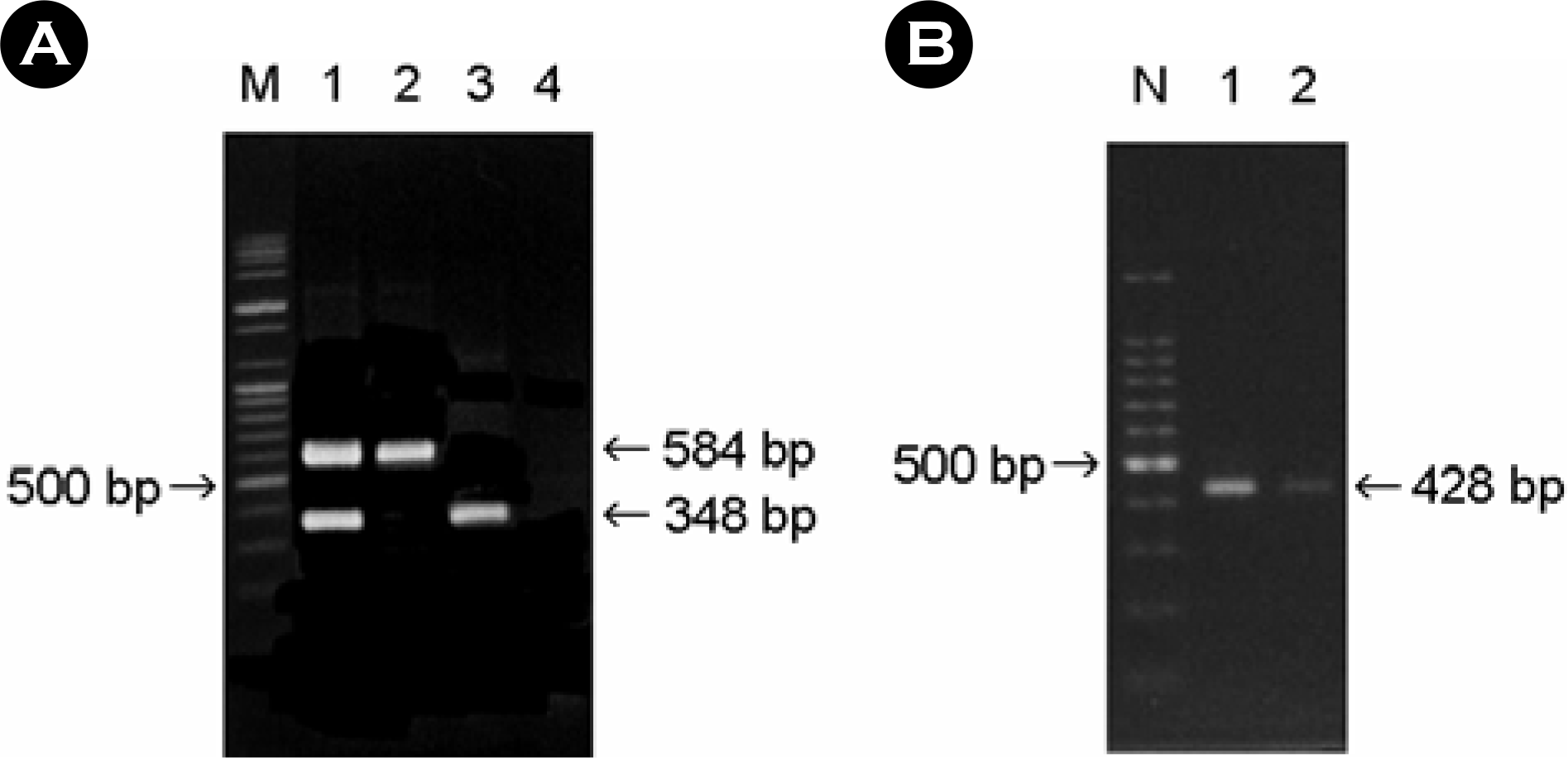
Figure 2.
Multiplex PCR to distinguish an intact selC locus from one disrupted by LEE. Primers K255 and K260 are predicted to produce a 418 bp amplicon in strains containing a LEE-disrupted locus. Primers K261 and K260 produced the 527 bp amplicon in strains indicating an intact selC locus. Lane M, 100 bp plus ladder (molecular weight markers); lane 1, E. coli EDL933 (stx1+, stx2+, control strains); lane 2, EHEC strain (O111:H–).
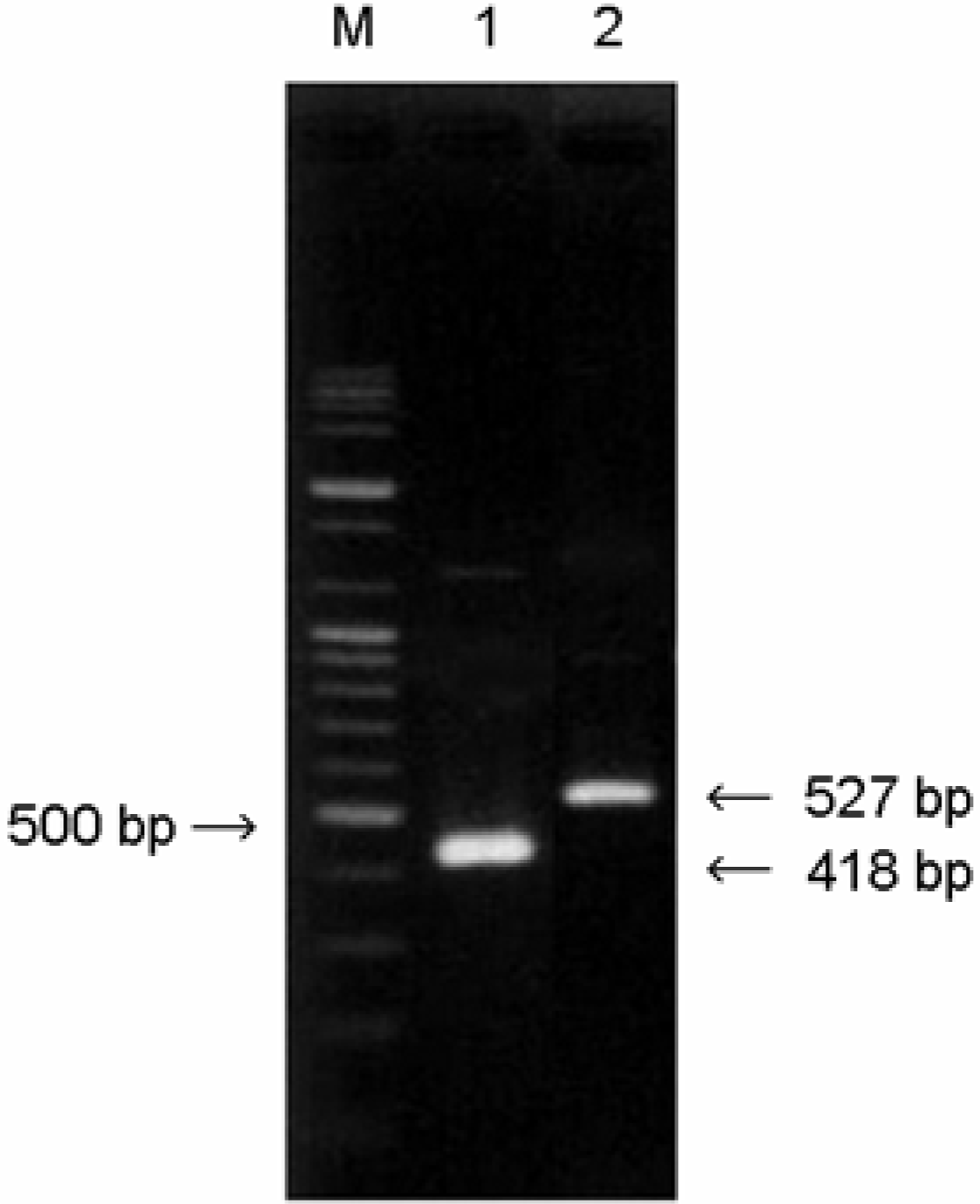
Table 1.
E. coli strains tested in this study and their sources
Table 2.
PCR primers used in the study
Table 3.
PCR primers for the detection of LEE-related genes in AEEC
| Genes and subtypes | Strains (accession number) | Sequences of the deduced primersa (5′-3′) | Annealing temperature (°C) | Amplicon (bp) |
|---|---|---|---|---|
| Disrupted selC | E2348/69 (GenBank AF031372) | K260: GAGCGAATATTCCGATATACTGGTT | 50.0 | 418 |
| K255: GGTTGAGTCGATTGATCTCTGG | ||||
| Intact selC | E2348/69 (GenBank AF031372) | K260: GAGCGAATATTCCGATATACTGGTT | 50.0 | 527 |
| K261: CCTGCAAATAAACACGGCGCAT | ||||
| eae type α | E2348/69 (EMBL X60439) | B73: TACTGAGATTAAGGCTGATAG | 50.4 | 452 |
| B138: GACCAGAAGAAGATCCG | ||||
| eae type β | EDL933 (EMBL Z11541) | B73: TACTGAGATTAAGGCTGATAG | 50.2 | 778 |
| B74: AGGAAGAGGGTTTTGTGT | ||||
| eae type γ | 193 (GenBank AF043226) | B73: TACTGAGATTAAGGCTGATAG | 50.8 | 520 |
| B137: TGTATGTCGCACTCTGAT | ||||
| tir type α | E2348/69 (GenBank AF022236) | B139: CRCCKCCAYTACCTTCACG | 54.2 | 342 |
| B152: CGCTAACCTCCAAACCAT | ||||
| tir type β | EDL933 (GenBank AF07134) | B139: CRCCKCCAYTACCTTCACG | 54.7 | 781 |
| B141: GTCGGCAGTTTCAGTTTCAT | ||||
| tir type γ | 95ZG1 (GenBank AF070068) | B139: CRCCKCCAYTACCTTCACG | 53.4 | 560 |
| B140: GATTTTTCCCTCGCCACTG | ||||
| espA type α | E2348/69 (GenBank Z54352) | B163: TGAGGCATCTAARGMGTT | 48.9 | 269 |
| B165: GCTGGCTATTATTGACC | ||||
| espA type β | EDL933 (GenBank Y13068) | B163: TGAGGCATCTAARGMGTT | 47.9 | 172 |
| B164: ATCACGAATACCAGTTACCG | ||||
| espA type γ | RDEC-1 (GenBank U80908) | B163: TGAGGCATCTAARGMGTT | 46.4 | 101 |
| B166: TGCCTTTCTTATTCTTGTCG | ||||
| espB type α | E2348/69 (GenBank AF022236) | B148: GCCGTTTTTGAGAGCCG | 50.6 | 94 |
| B151: TCCCCAGGACAGATGAGA | ||||
| espB type β | EDL933 (GenBank Y13068) | B148: GCCGTTTTTGAGAGCCG | 53.1 | 188 |
| B150: GCACCAGCAGCCTTTGG | ||||
| espB type γ | RDEC-1 (GenBank U80796) | B148: GCCGTTTTTGAGAGCCG | 50.9 | 233 |
| B149: CTTTCCGTTGCCTTAG | ||||
| espD type α | E2348/69 (GenBank AF022236) | B186: CGAAGAACAACAAAAAGCC | 52.8 | 492 |
| B188: ACAGCAAAAGCAGAAACCT | ||||
| espD type β | E22 (GenBank AF054421) | B186: CGAAGAACAACAAAAAGCC | 53.4 | 414 |
| B187: GCAGAGGTCGTAAATCCAT | ||||
| espD type γ | EDL933 (GenBank AF071034) | B186: CGAAGAACAACAAAAAGCC | 53.8 | 350 |
| B189: CTGCCGCTTTCTCAACGAC |
Table 4.
Serotypes of AEEC used in this study
Table 5.
Presence and typing of LEE-related genes in AEEC strains
| Strains | Sources | selC disrupted by LEE | Pathotypea | No. of strains | Serotype |
|---|---|---|---|---|---|
| EHEC | PNUH | yes | eaeγ-tirγ-espAγ-espBγ-espDγ | 2 | O157:H7 |
| CSEAS | yes | eaeγ-tirγ-espAγ-espBγ-espDγ | 20 | O157:H7 | |
| KUH | yes | eaeγ-tirγ-espAγ-espBγ-espDγ | 16 | O157:H7, O157:H– | |
| yes | eaeα-tirα-espA–espBα-espD– | 2 | O157:H45 | ||
| yes | eaeβ-tirβ-espAβ-espBβ-espDβ | 2 | O26:H11 | ||
| no | eaeγ-tirα-espAα-espBα-espD– | 2 | O111:H– | ||
| EPEC | KUH | yes | eaeγ-tirγ-espAγ-espB–-espDγ | 6 | O55:H7 |
| no | ND | 3 | O55:H10 | ||
| yes | eaeα-tirα-espAα-espBα-espDα | 2 | O55:H6 | ||
| no | ND | 1 | O55:H– | ||
| yes | eaeγ-tirγ-espAγ-espB—espD– | 2 | O111:H12 | ||
| EHEC (EDL933) | Control strain | yes | eaeγ-tirγ-espAγ-espBγ-espDγ | 1 | O157:H7 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download