Abstract
It has been reported that most of Helicobacter pylori proteome components appear so crowded in the region of pH 4.5∼8.0 that a lot of them were inseparable in 2-DE using the broad range IPG strip. Therefore, inseparable protein spots in 2-DE profiles have to be apart from each other for improving the protein identification. Here, we attempt to examine the usability of the narrow range IPG strips for separating close spots in the broad range IPG strip at proteomic analysis of H. pylori. The whole cell proteins of H. pylori strain 26695 were separated by narrow range IPG strips (pI 3.9∼5.1, 4.7∼5.9, 5.5∼6.7, and 6.3∼8.3, respectively), followed by SDS-PAGE, and visualized by silver staining, showing that the distances between spots were widened and the total number of detectable spots was increased. Resolved protein spots were identified by the peptide fingerprinting using MALDI-TOF-MS. As a result, 87 expressed proteins were identified by the peptide fingerprinting. Of them, 23 proteins, including hydrogenase expression/formation protein, purine-binding chemotaxis protein, and ribosomal protein S6, have not been reported in the previous proteome studies of H. pylori. Thus, these results demonstrate that the high complexity proteome components could be effectively separated using the narrow range IPG strips, which might be helpful to strengthen the contents of the master protein map of the H. pylori reference strain.
References
1). Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 14:176–180. 1999.
2). Backert S, Kwok T, Schmid M, Selbach M, Moese S, Peek RM Jr, Konig W, Meyer TF, Jungblut PR. Subproteomes of soluble and structure-bound Helicobacter pylori proteins analyzed by two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 5:1331–1345. 2005.
3). Baik SC, Kim JB, Cho MJ, Kim YC, Park CG, Rhou HH, Choi HJ, Rhee KH. Prevalence of Helicobacter pylori infection among normal Korean adults. J Korean Soc Microbiol. 6:455–462. 1990.
4). Baik SC, Kim KM, Song SM, Kim DS, Jun JS, Lee SG, Song JY, Park JU, Kang HL, Lee WK, Cho MJ, Youn HS, Ko GH, Rhee KH. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of Helicobacter pylori strain 26695. J Bacteriol. 186:949–955. 2004.
5). Baik SC, Yoon HS, Chung MH, Lee WK, Cho MJ, Ko KH, Park CK, Kasai H, Rhee KH. Increased oxidative DNA damage Helicobacter pylori-infected human gastric mucosa. Cancer Res. 56:1279–1282. 1996.
6). Banatvala NK, Mayo K, Megraud F, Jennings R, Deeks JJ, Feldman RA. The cohort effect and Helicobacter pylori. J Infect Dis. 168:219–221. 1993.
7). Blaser MJ. Gastric Campylobacter-like organisms gastritis and peptic ulcer disease. Gastroenterology. 97:371–383. 1987.
8). Blaser MJ. Helicobacter pylori and gastric diseases. Br Med J. 316:1507–1510. 1998.
10). Cho MJ, Jeon BS, Park JW, Jung TS, Song JY, Lee WK, Choi YJ, Choi SH, Park SG, Park JU, Choe MY, Jung SA, Byun EY, Baik SC, Youn HS, Ko GH, Lim D, Rhee KH. Identifying the major proteome components of Helicobacter pylori strain 26695. Electrophoresis. 23:1161–1173. 2002.
11). Dunn BE, Cohen H, Blaser MJ. Helicobacter pylori. Clin Microbiol Rev. 10:720–741. 1997.
12). Fischer W, Haas R. The RecA protein of Helicobacter pylori requires a posttranslational modification for full activity. J Bacteriol. 186:777–784. 2004.
13). Forman D, Newell DG, Fullerton F, Yarnel JW, Stacey AR, Wald N, Sitas F. Association between infection with Helicobacter pylori and risk of gastric cancer: evidence from a prospective investigation. Br Med J. 302:1302–1305. 1991.
14). Gharahdaghi F, Weinberg CR, Maagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis. 20:601–605. 1999.

15). Görg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, Weiss W. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 21:1037–1053. 2000.

16). Görg A, Obermaier C, Boguth G, Weiss W. Recent developments in two-dimensional gel electrophoresis with immobilized pH gradients: wide pH gradients up to pH 12, longer separation distances and simplified procedures. Electrophoresis. 20:712–717. 1999.

17). Humphery-Smith I, Cordwell SJ, Blackstock WP. Proteome research: complementarity and limitations with respect to the RNA and DNA worlds. Electrophoresis. 18:1217–1242. 1997.

18). Jungblut PR, Bumann D, Haas G, Zimmy-Arndt U, Holland P, Lamer S, Siejak F, Aebischer A, Meyer TF. Comparative proteome analysis of Helicobacter pylori. Mol Microbiol. 36:710–725. 2000.
19). Kim H, Wu CA, Kim DY, Han YH, Ha SC, Kim CS, Suh SW, Kim KK. Crystallization and preliminary X-ray crystallographic study of UDP-glucose pyrophosphorylase (UGPase) from Helicobacter pylori. Acta Crystallogr D Biol Crystallogr. 60:1447–1449. 2004.
20). Lee HW, Choe YH, Kim DK, Jung SY, Lee NG. Proteomic analysis of a ferric uptake regulator mutant of Helicobacter pylori: regulation of Helicobacter pylori gene expression by ferric uptake regulator and iron. Proteomics. 4:2014–2027. 2004.
21). Lock RA, Cordwell SJ, Coombs GW, Walsh BJ, Forbes GM. Proteome analysis of Helicobacter pylori: major proteins of type strain NCTC11637. Pathol. 33:365–374. 2001.
22). Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 16:1311–115. 1984.

23). McAtee CP, Hoffman PS, Berg DE. Identification of differentially regulated proteins in metronidozole resistant Helicobacter pylori by proteome techniques. Proteomics. 1:516–521. 2001.
24). Nomura A, Stemmermann GN, Chyou PH, Kato I, Perez-Perez GI, Blaser J. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 325:1132–1136. 1991.
25). O'Connell KL, Stults JT. Identification of mouse liver proteins on two-dimensional electrophoresis gels by matrix-assisted laser desorption/ionization mass spectrometry of in situ enzymatic digests. Electrophoresis. 18:349–359. 1997.
26). Oh JD, Kling-Bäckhed H, Giannakis M, Xu J, Fulton RS, Fulton LA, Cordum HS, Wang C, Elliott G, Edwards J, Mardis ER, Engstrand LG, Gordon JI. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc Natl Acad Sci U S A. 103:9999–10004. 2006.
27). Parsonnet J, Hansen S, Rodriguez L, Gelb AB, Warnke RA, Jellum E, Orentreich N, Vogelman JH, Friedman GD. Helicobacter pylori infection and lymphoma. N Engl J Med. 330:1132–1136. 1994.
28). Peterson W. Helicobacter pylori and peptic ulcer disease. N Engl J Med. 324:1043–1048. 1991.
29). Proteome 2D-PAGE database. http://www.mpiib-berlin.mpg.de/2D-PAGE/.
31). Rhee KH, Yoon HS, Baik SC, Lee WK, Cho MJ, Choi HJ, Maeng KY, Ko KW. Prevalence of Helicobacter pylori infection among normal Koreans. J Korean Soc Microbiol. 6:475–490. 1990.
32). Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 7:539–547. 1997.
33). Uwins C, Deitrich C, Argo E, Stewart E, Davidson I, Cash P. Growth-induced changes in the proteome of Helicobacter pylori. Electrophoresis. 27:1136–1146. 2006.
34). Wildgruber R, Harder A, Obermaier C, Boguth G, Weiss W, Fey SJ, Larsen PM, Görg A. Towards higher resolution: two-dimensional electrophoresis of Saccharomyces cerevisiae proteins using overlapping narrow immobilized pH gradients. Electrophoresis. 21:2610–2616. 2000.
Figure 1.
Two-dimensional-gel electrophoresis of whole cell proteins from H. pylori strain 26695. Whole cell proteins solution was separated on a narrow range IPG strips, followed by 12.5% SDS-PAGE, and visualized by silver staining. The original gel size was 20×18×0.15 cm. (A) pH 3.9∼5.1; (B) pH 4.7∼5.9; (C) pH 5.5∼6.7; (D) pH 6.3∼8.3.
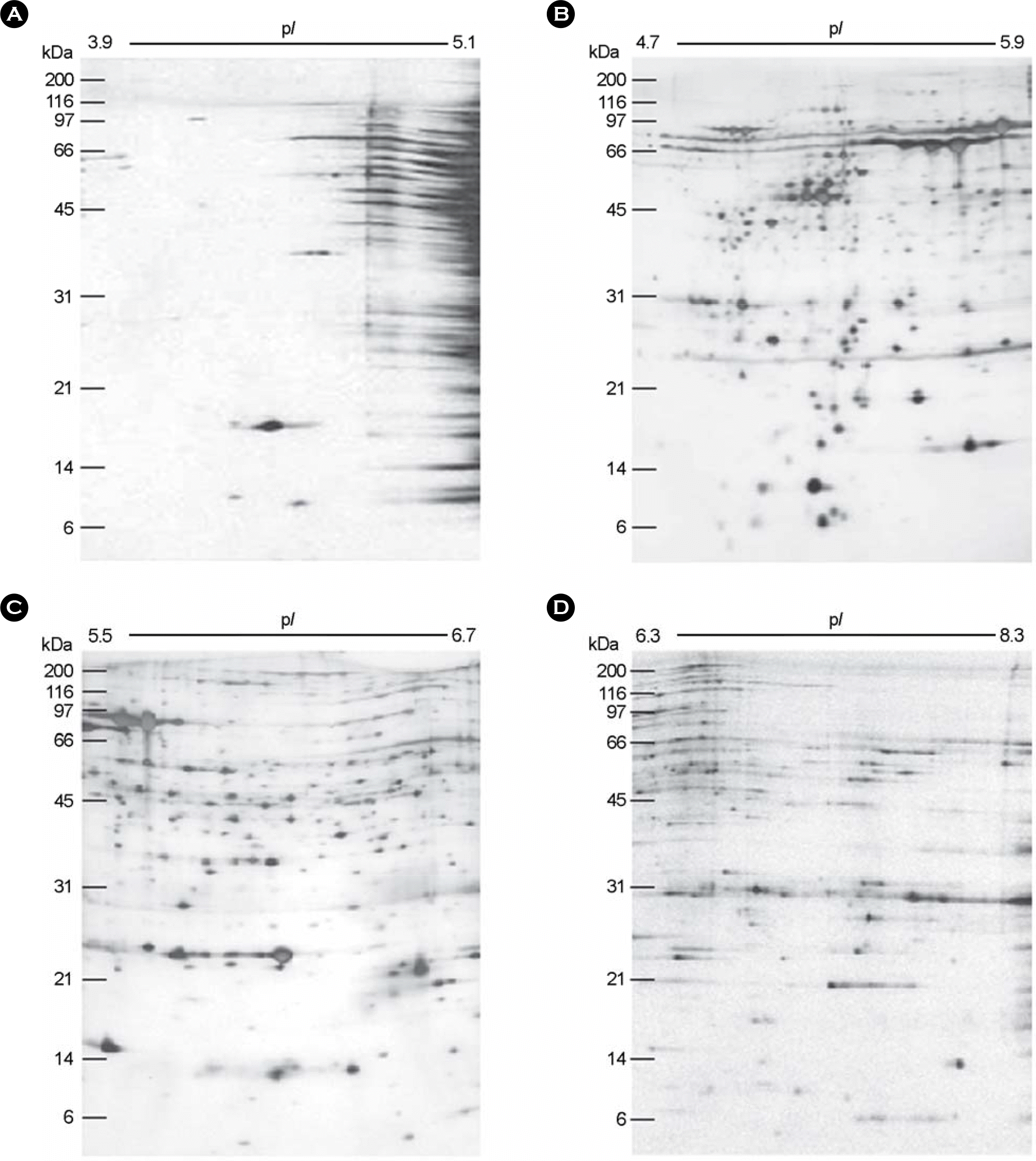
Figure 2.
The composite profile of the whole cell proteins of H. pylori strain 26695 resolved in the 2-DE using the narrow pH range IPG strips. The whole cell proteins of H. pylori strain 26695 was subjected to 2-DE using IPG strips of 3.9∼5.1, 4.7∼5.9, 5.5∼6.7, and 6.3∼8.3 ranges, the gels were visualized with silver staining. The 4 kinds of images were taken up and analyzed with PDQUEST software, and excluded with the overlapping zones of each right and left termini, and then combined by PHOTOSHOP software to generate the composite one ranged from pI 3.9 to pI 8.3. The marked protein spots have been identified and listed in Table 1.
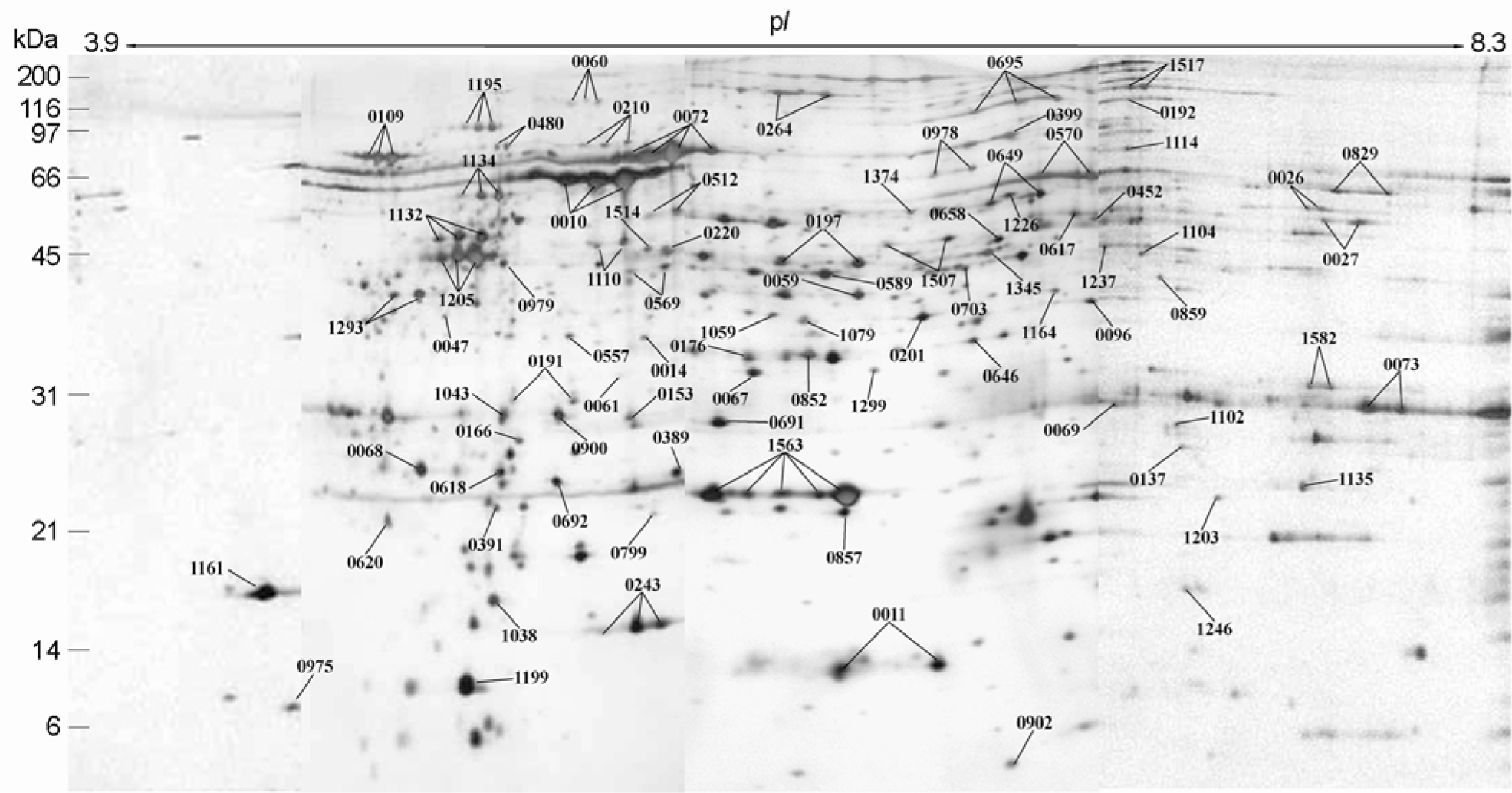
Figure 3.
Blow-up images of two different regions in 2-DE gels of H. pylori whole cell proteins resolved by broad (pH 3.0∼10.0), intermediate (pH 5.0∼8.0), narrow (pH 4.7∼5.9) pH strips. Here, the narrower is the pH range of the strip, the higher resolution does the spots profile show in which the resolution and the number of spots are improved. HtpG spot appears in the gel of narrow pH strips but did not in otherwise.
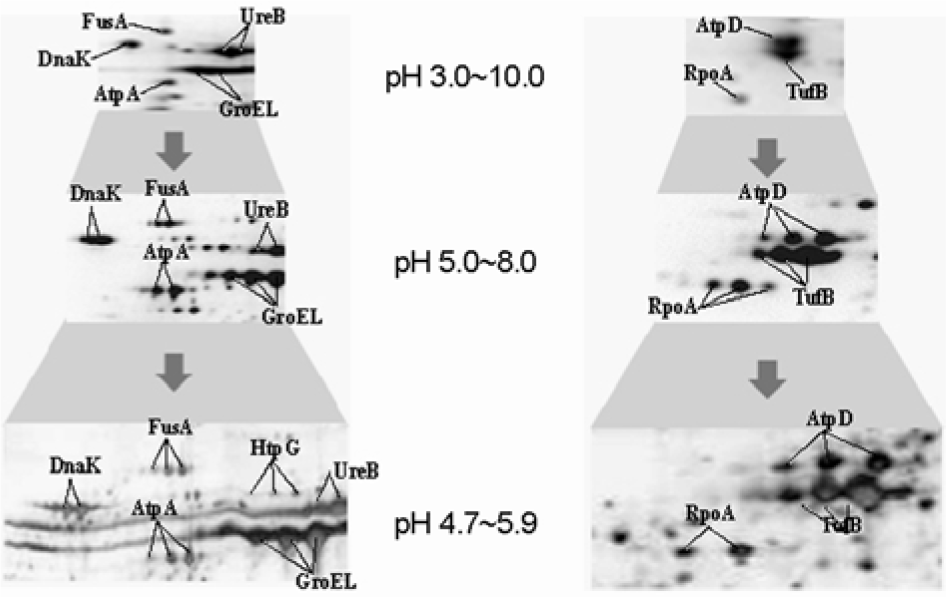
Table 1.
Protein identification of spots from narrow range 2-DE gels the whole cell proteins of H. pylori strain 26695
| TIGR locus§ | MW (Da) | pI | Annotation |
|---|---|---|---|
| HP 0010 | 58263.91 | 5.4772 | Chaperone and heat shock protein (groEL) |
| HP 0011 | 12990.87 | 6.5917 | Co-chaperone (groES) |
| HP 0014 | 31681.02 | 5.7841 | Hypothetical protein |
| HP 0026 | 48350.02 | 8.0209 | Citrate synthase (gltA) |
| HP 0027 | 47531.05 | 7.9527 | Isocitrate dehydrogenase (icd) |
| HP 0047∗ | 36405.28 | 4.8329 | Hydrogenase expression/formation protein (hypE) |
| HP 0059∗ | 32723.51 | 4.8431 | Hypothetical protein |
| HP 0060∗ | 93128.96 | 4.8450 | Hypothetical protein |
| HP 0061∗ | 21764.88 | 5.9064 | Hypothetical protein |
| HP 0067 | 29722.06 | 6.1629 | Urease accessory protein (ureH) |
| HP 0068 | 21955.25 | 4.7602 | Urease accessory protein (ureG) |
| HP 0069 | 28618.77 | 7.6498 | Urease accessory protein (ureF) |
| HP 0072 | 61683.52 | 5.9110 | Urease beta subunit (ureB) |
| HP 0073 | 26539.49 | 8.9593 | Urease, alpha subunit (ureA) |
| HP 0096 | 34796.18 | 7.0468 | Phosphoglycerate dehydrogenase |
| HP 0109 | 67051.91 | 4.7568 | Chaperone and heat shock protein70 (dnaK) |
| HP 0137∗ | 23610.48 | 7.6666 | Hypothetical protein |
| HP 0153∗ | 37686.28 | 5.5065 | Recombinase (recA) |
| HP 0166 | 25855.46 | 5.0781 | Response regulator (ompR) |
| HP 0176 | 33772.69 | 6.2517 | Fructose-bisphosphate aldolase (tsr) |
| HP 0191 | 27651.76 | 5.1972 | Fumarate reductase, iron-sulfur subunit (frdB) |
| HP 0192 | 80121.10 | 7.3217 | Fumarate reductase, flavoprotein subunit (frdA) |
| HP 0197 | 42362.06 | 6.4292 | S-adenosylmethionine synthetase 2 (metX) |
| HP 0201 | 36483.11 | 6.5193 | Fatty acid/phospholipid synthesis protein (plsX) |
| HP 0210 | 71274.03 | 5.2608 | Chaperone and heat shock protein C62.5 (htpG) |
| HP 0220 | 42403.38 | 6.1542 | Synthesis of [Fe-S] cluster (nifS) |
| HP 0243 | 16933.34 | 5.8309 | Neutrophil activating protein (napA) (bacterioferritin) |
| HP 0264 | 96683.36 | 6.2660 | ATP-dependent protease binding subunit (clpB) |
| HP 0389 | 24617.63 | 6.1683 | Superoxide dismutase (sodB) |
| HP 0391∗ | 18966.68 | 5.0808 | Purine-binding chemotaxis protein |
| HP 0399 | 62826.43 | 6.7727 | Ribosomal protein S1 (rps1) |
| HP 0452∗ | 60506.79 | 5.9621 | Conserved hypothetical protein |
| HP 0480 | 66676.31 | 5.0937 | GTP-binding protein, fusA-homolog (yihK) |
| HP 0512 | 54513.58 | 6.0379 | Glutamine synthetase (glnA) |
| HP 0557 | 34881.13 | 5.3110 | Acetyl-coenzyme A carboxylase (accA) |
| HP 0569 | 40574.45 | 5.5302 | GTP-binding protein (gtp1) |
| HP 0570 | 54433.07 | 7.1054 | Aminopeptidase a/i (pepA) |
| HP 0589 | 41508.81 | 6.3623 | Ferredoxin oxidoreductase, alpha subunit |
| HP 0617 | 65601.53 | 6.8834 | Aspartyl-tRNA synthetase (aspS) |
| HP 0618 | 21243.21 | 5.0039 | Adenylate kinase (adk) |
| HP 0620 | 19271.89 | 4.8028 | Inorganic pyrophosphatase (ppa) |
| HP 0646∗ | 30975.69 | 6.5192 | UDP-glucose pyrophosphorylase (galU) |
| HP 0649 | 51980.86 | 6.8341 | Aspartate ammonia-lyase (aspA) |
| HP 0658 | 53288.14 | 6.0094 | Glutamyl-tRNA(Gln) amidotransferase subunit B (gatB) |
| HP 0691 | 25362.24 | 5.8082 | 3-oxoadipate coA-transferase subunit A (yxjD) |
| HP 0692 | 22262.85 | 5.3450 | 3-oxoadipate coA-transferase subunit B (yxjE) |
| HP 0695 | 78532.79 | 6.9435 | Hydantoin utilization protein A (hyuA) |
| HP 0703 | 43414.34 | 6.7535 | Response regulator |
| HP 0799∗ | 19676.86 | 5.3363 | Molybdopterin biosynthesis protein (mog) |
| HP 0829 | 51801.80 | 8.2155 | Inosine-5′-monophosphate dehydrogenase (guaB) |
| HP 0852∗ | 35776.74 | 6.6806 | Hypothetical protein |
| HP 0857∗ | 21102.26 | 6.3967 | Phosphoheptose isomerase (gmhA) |
| HP 0859 | 37412.66 | 7.3588 | ADP-L-glycero-D-mannoheptose-6-epimerase (rfaD) |
| HP 0900 | 27310.33 | 5.2449 | Hydrogenase expression/formation protein (hypB) |
| HP 0902 | 11029.96 | 6.6745 | Hypothetical protein |
| HP 0975∗ | 10656.15 | 4.7389 | Glu-tRNA(Gln) amidotransferase, subunit C |
| HP 0978 | 54492.59 | 6.6324 | Cell division protein (ftsA) protein |
| HP 0979∗ | 40954.34 | 5.1659 | Cell division protein (ftsZ) |
| HP 1038 | 18483.25 | 4.9294 | 3-dehydroquinase type II (aroQ) |
| HP 1043∗ | 25468.16 | 4.9740 | Response regulator |
| HP 1059 | 37360.31 | 6.0818 | Holliday junction DNA helicase (ruvB) |
| HP 1079∗ | 42919.70 | 6.3237 | Hypothetical protein |
| HP 1102 | 25727.47 | 7.3447 | Glucose-6-phosphate 1-dehydrogenase (devB) |
| HP 1104 | 38645.51 | 7.4067 | Cinnamyl-alcohol dehydrogenase ELI3–2 (cad) |
| HP 1110 | 44743.62 | 5.8122 | Pyruvate ferredoxin oxidoreductase, alpha subunit |
| HP 1114 | 75916.20 | 6.9096 | Excinuclease ABC subunit B (uvrB) |
| HP 1132 | 51478.08 | 5.0767 | ATP synthase F1, subunit beta (atpD) |
| HP 1134 | 55143.11 | 5.0512 | ATP synthase F1, subunit alpha (atpA) |
| HP 1135∗ | 20368.81 | 8.4730 | ATP synthase F1, subunit delta (atpH) |
| HP 1161 | 17492.37 | 4.2040 | Flavodoxin (fldA) |
| HP 1164 | 35985.98 | 6.8739 | Thioredoxin reductase (trxB) |
| HP 1195 | 77020.88 | 5.0111 | Translation elongation factor EF-G (fusA) |
| HP 1199 | 13313.38 | 4.9125 | Ribosomal protein L7/L12 (rpl7/l12) |
| HP 1203 | 20261.24 | 7.8735 | Transcription termination factor NusG (nusG) |
| HP 1205 | 43647.70 | 4.9302 | Translation elongation factor EF-Tu (tufB) |
| HP 1226∗ | 40196.14 | 6.5108 | Oxygen-independent coproporphyrinogen III oxidase (hemN) |
| HP 1237∗ | 41635.41 | 7.1579 | Carbamoyl-phosphate synthetase (pyrAa) |
| HP 1246∗ | 16971.34 | 7.4983 | Ribosomal protein S6 (rps6) |
| HP 1293 | 38499.11 | 4.6887 | DNA-directed RNA polymerase, alpha subunit (rpoA) |
| HP 1299 | 27576.93 | 6.4208 | Methionine amino peptidase (map) |
| HP 1345∗ | 44771.64 | 6.7477 | Phosphoglycerate kinase |
| HP 1374∗ | 50353.20 | 6.4377 | ATP-dependent protease ATPase subunit (clpX) |
| HP 1507∗ | 43904.36 | 6.5698 | Conserved hypothetical ATP-binding protein |
| HP 1514 | 44649.63 | 5.5915 | Transcription termination factor NusA (nusA) |
| HP 1517 | 149715.58 | 7.5020 | Type IIS restriction enzyme R and M protein (ECO57IR) |
| HP 1563 | 22235.57 | 6.2482 | Alkyl hydroperoxide reductase (tsaA) |
| HP 1582 | 29720.44 | 8.0532 | Pyridoxal phosphate biosynthetic protein J (pdxJ) |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download