Abstract
According to the serological screening methods of antigen-antibody reaction such as ELISA, it has been known that the complete detection of viral infections of HBV, HCV, and HIV-1 viruses in the blood and blood related-products is not much reliable. Therefore, nucleic acid amplification testing methods (NAT) adopted to detect the small quantitative viral nucleic acids could support the basis of using and supplying the blood and its related products safely. This research work is basically designed to describe the simultaneous blood screening system by multiplex or duplex tests for detection of HBV, HCV, and HIV-1 viruses in the blood at one time with low price and labor. It is aimed at easy detection by using the conventional agarose gel electrophoresis. Thus, we tried to detect and identify the viral components in the blood sample according to their different size of PCR products. We decided a set of consensus sequences to recognize each viral DNA fragments after running the multiplex PCR in one tube. This was done by nested RT-PCR using two different RNA viral genomic templates followed by multiplex PCR with addition of viral DNA and their primers after purifying the viral genomic nucleic acids. Those specific primers could be used without any interference to amplify each viral genome in the blood samples. The sensitivities with different viral loads were evaluated on the agarose gel electrophoresis. Three different viral agents in the blood samples could be tested by this multiplex (RT)-PCR with three different primers.
References
1). Allain J-P, Hewitt PE, Tedder RS, Williamson LM. Evidence that anti-HBc but not HBV DNA testing may prevent some HBV transmission by transfusion. Br J Hematol. 107:186–195. 1999.

2). Biswas R, Tabor E, Hsia CC, Wright DJ, Laycock MF, Fiebig EW, Peddada I, Smith R, Schreiber GB, Epstein JS, Nemo GJ, Busch MP. Comparative sensitivity of HBV NATs and HBsAg assays for detection of acute HBV infection. Transfusion. 43:788–798. 2003.

3). Candotti D, Adu-Darkodie Y, Davies F, Baldrich-Rubio E, Strirups F, Lee H, Allain J-P. AIDS in an HIV-seronegative Ghanaian women with intersubtype A/G recombinant HIV-1 infection. J Med Viro. 62:1–8. 2000.
4). Candotti D, Sarkodie F, Allain J-P. Frequent recovery and broad genotype 2 diversity characterize HCV infection in Ghana, West Africa. J Virol. 77:7914–7923. 2003.
5). Candotti D, Temple J, Owusu-Ofori S, Allain J-P. Multiplex real-time quantitative RT-PCR assay for hepatitis B virus, hepatitis C virus, and human immunodeficiency virus type 1. J of Virol Methods. 118:39–47. 2004.

6). Cuypers HTM, van Dijk R, Viret J-P, Schottstedt V, Lankinen M, Da Silba Cardoso M, Lelie PN. European multi-centre validation study of NucliSens Extractor in combination with HCV Amplicor 2.0 assay for HCV-NAT screening of plasma pools. Biologicals. 27:303–314. 1999.

7). Defoort J.-P, Marin M, Casano M, Prato S, Camilla C, Fert V. Simultaneous detection of multiplex-amplified HIV-1 RNA, HCV RNA, and HBV DNA using a flow cytometer microsphere-based hybridization assay. J Clinical Microbiol. 38:1066–1071. 2000.
8). Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE. Mutiplex PCR oprimization and application in diagnostic virology. Clinical Microbiol Rev. 13:559–570. 2000.
9). FDA. Guideline for the manufacture of in vitro diagnostic products. 1994.
10). Giachetti C, Linnen JM, Kolk DP, Dockter J, Gillotte-Tayler K, Park M, Ho M, McCormick MK, Minnes LT, McDonough ST. Highly sensitive multiplex assay for detection of HIV-1 and HCV RNA. J Clinical Microbiol. 40:2408–2419. 2002.
11). Gobbers E, Oosterlaken TAM, van Bussel MJA, Melsert R, Kroes ACM, Class ECJ. Efficient extraction of virus DNA by Nuclisens extractor allows sensitive detection of HBV virus by PCR. J of Clinical Microbiol. 39(12):4339–43. 2001.
12). Jackson BR, Busch MP, Stramer SL, AuBuchon JP. The cost-effectiveness of NAT for HIV, HCV, and HBV in whole blood donations. Transfusion. 43:721–729. 2003.
13). Kolk DP, Dockter J, Linnen J, Ho-Sing-Loy M, Gillotte-Taylor K, McDonough SH, Mimms L, Giachetti C. Significant closure of the human immunodeficiency virus type 1 and hepatitis C virus preseroconversion detection windows with a transcription-mediated-amplification-driven assay. J Clin Microbiol. 40(5):1761–6. 2002.

14). Linnen J, Broulik A, Umali A, Kolk D, Dockter J, McDonogh S, Mimms L, Giachetti C. Analytical sensitivity of the TMA-based Procleix Ultrio assay and effect of assay sensitivity on closing the HBV detection window. Transfusion. 42:8S. 2002.
15). Meng Q, Wong C, Rangachari A, Tamatsukuri S, Sasaki M, Fiss E, Cheng L, Ramankutty T, Clarke D, Yawata H, Sakakura Y, Hirose T, Impraim C. Automated Multiplex assay system for simultaneous detection of HBV DNA, HCV RNA, HIV-1 RNA. J of Clinical Microbiol. 39(8):2937–45. 2001.
16). Mercier B, Burlot L, Ferec C. Simultaneous screening for HBV DNA and HCV RNA genomes in blood donations using a novel TaqMan PCR assay. J Virol Methods. 77:1–9. 1999.

17). Mine H, Emura H, Miyamoto M, Tomono T, Minegishi K, Murokaa H, Yamanak R, Yoshikawa A, Nishioka K. Japanese red Cross NAT Research Group. High throughput screening of 16 million serologically negative blood donors for HBV, HCV, HIV-1 by NAT testing with specific and sensitive multiplex reagent in Japan. J of Virol Methods. 112:145–151.
18). Miyachi H, Masukawa A, Ohshima T, Hirose T, Impraim C, Ando Y. Automated specific capture of HCV RNA with probes and paramagnetic separation. J of Clinical Microbiol. 38:18–21. 2003,. 2000.
19). Ohnuma H, Tanaka T, Yoshikawa A, Murokawa H, Minegishi K, Yamanaka R, Lizuka HY, Miyamoto M, Satoh S, Nakahira S, Tomono T, Murozuka T, Takeda Y, Doi Y, Mine H, Yokoyama S, Hirose T, Nishioka K. Japanese Red Cross NAT Screening Research Group. The first large-scale nucleic acid amplification testing (NAT) of donated blood using multiplex reagent for simultaneous detection of HBV, HCV, and HIV-1 and significance of NAT for HBV. Microbial Immunol. 45(9):667–672. 2001.

20). Polz MF, Cavanaugh CM. Bias in template-to product ratios in multitemplate PCR. Appl Environ Microbiol. 64:3724–3730. 1998.
21). Roth WK, Weber M, Seifried E. Feasibility and efficacy of routine PCR screening of blood donations for HCV, HBV, and HIV-1 in a blood-bank setting. Lancet. 353:359–363. 1999.
22). Schreiber GB, Busch MP, Kleinman SH, Korelitz JJ. The risk of transfusion-transmitted viral infection. The retrovirus epidemiology donor study. New Eng J Med. 334:1685–1690. 1996.
23). Soldan K, Barbara JA, Ramsey ME, Hall AJ. Estimation of the risk of HBV, HCV and HIV infectious donations entering the blood supply in England. Vox Sang. 84:274–286. 2003.
24). Stramer SL, Waldman KT, Brodsky P, Davis JT. Comparison of HIV-1/HCV nucleic acid test reactivities of diluted and undiluted donations. Transfusion. 40:S94–040A. 2000.
25). Vet JA, Majithia AR, Marras SAE, Tyagi S, Dube S, Poiesz BJ, Kramer FR. Multiplex detection of four pathogenic retro-viruses using molecular beacons. PNAS USA. 96:6394–6399. 1999.

26). Weinberger KM, Widenmann E, Bohm S, Jilg W. Sensitive and accurate quantitation of HBV DNA using kinetic fluorescence detection system (TaqMan PCR). J Virol Methods. 85:75–82. 2000.
27). Widel A, Edmund H, Persson MH, Jonsson M. Transmission of HCV via both erythrocyte and platelet transfusions from a single donor in serological window-phase of HCV. Vox Sang. 71:55–57. 1966.
28). Yates S, Penning M, Goudsmit J, Frantzen I, Weije B, Strijp D, Gemen B. Quantitative detection of HBV DNA by real-time NA sequence-based amplification with molecular Beacon detection. J of Clinical Microbiol. 39:3656–3665. 2001.
Figure 1.
Detection limit of HBV (A), HCV (B), HIV-1 (C) DNA plasmid samples using PCR. M, 100 bp DNA ladder; Each lanes was half-diluted from 240 ng/ml (A), 240 ng/ml (B), 1,300 ng/ml (C). Positive PCR product were shown on the 336 bp size (A), 250 bp size (B), and 501 bp size (C), respectively.
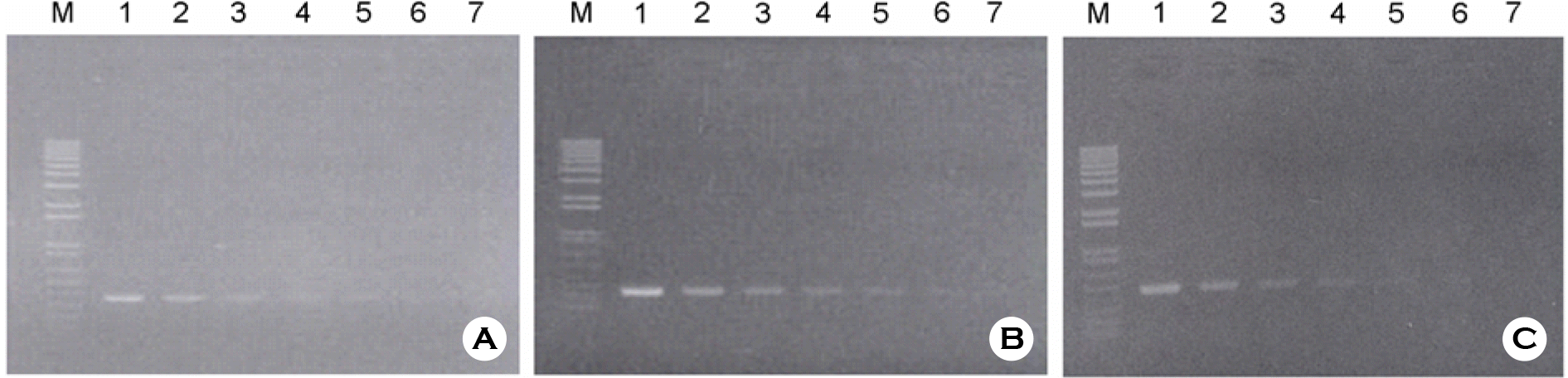
Figure 2.
(A) Detection limit of Multiplex PCR with combination of different plasmid NAs of HBV, HCV, and HIV-1 samples on the 1.4% agarose gel: at the lane 1, HBV (60 ng/ml) plus HCV (45 ng/ml) plus HIV-1 (325 ng/ml) were mixed with 1 μl of each into the reaction volume, and lane 2 was used with half of lane 1, lane 3 was used with half of lane 2, and so on; (B) Detection limit of multiplex PCR with labeled with 32P-dTTP was carried on the 5% Polyacrylamide gel: C, positive control; lane 1, 0.1 μg/ml of each plasmid DNAs, respectively; lanes 2∼8 were used with serial dilution of 10 fold, respectively. The upper bands are shown as HIV-specific DNA (501 bp), middle bands are shown as HBV-specific DNA (336 bp), and lower bands are shown as HCV-specific DNA (250 bp).
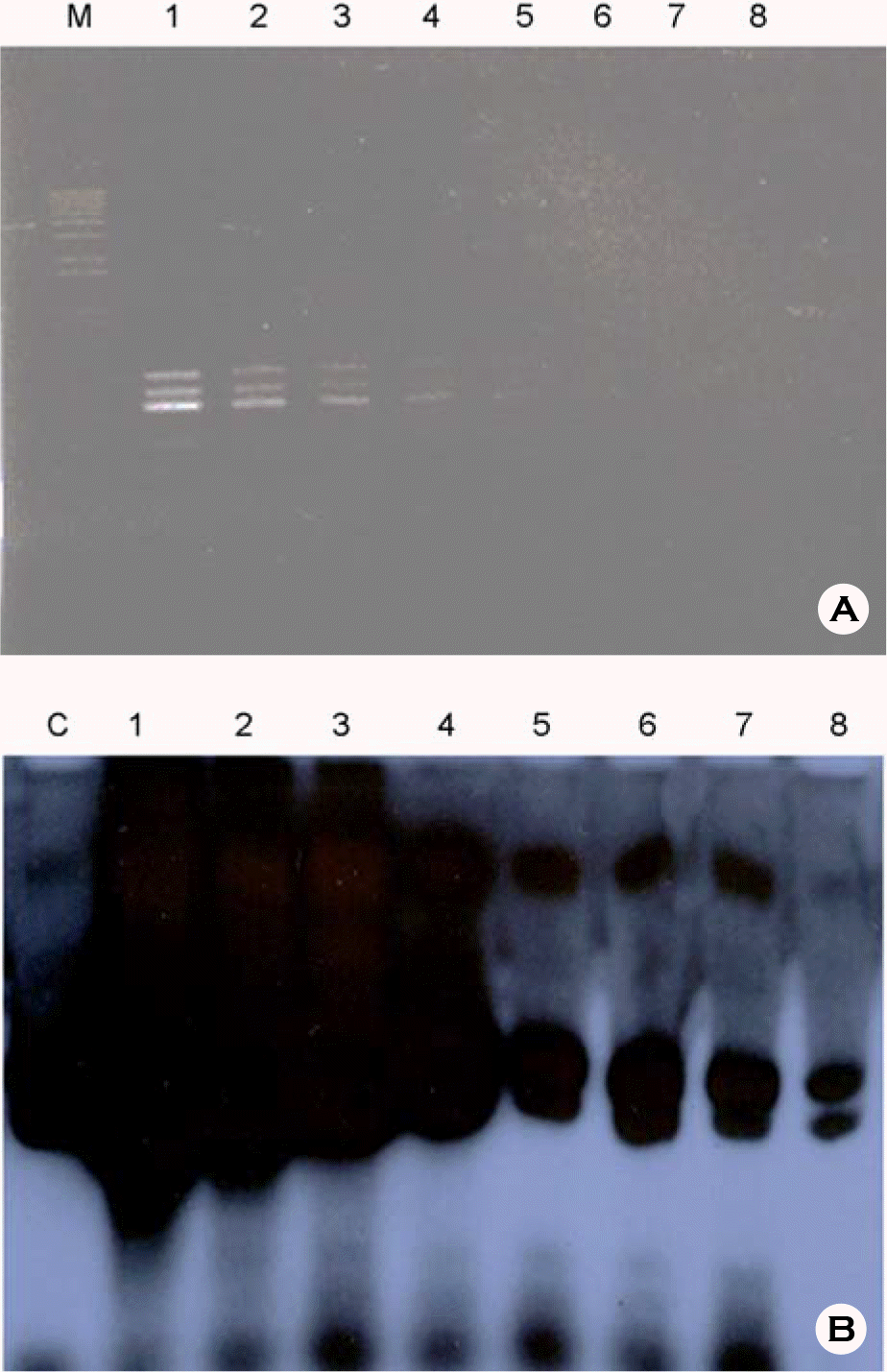
Figure 3.
Reproducibility of (RT)-PCR methods HBV (A), HCV (B), HIV (C) DNA PCRs were performed three times by three different persons, respectively. 1, higher concentration than detection limit; 2, similar concentration of detection limit; 3, lower concentration than detection limit.
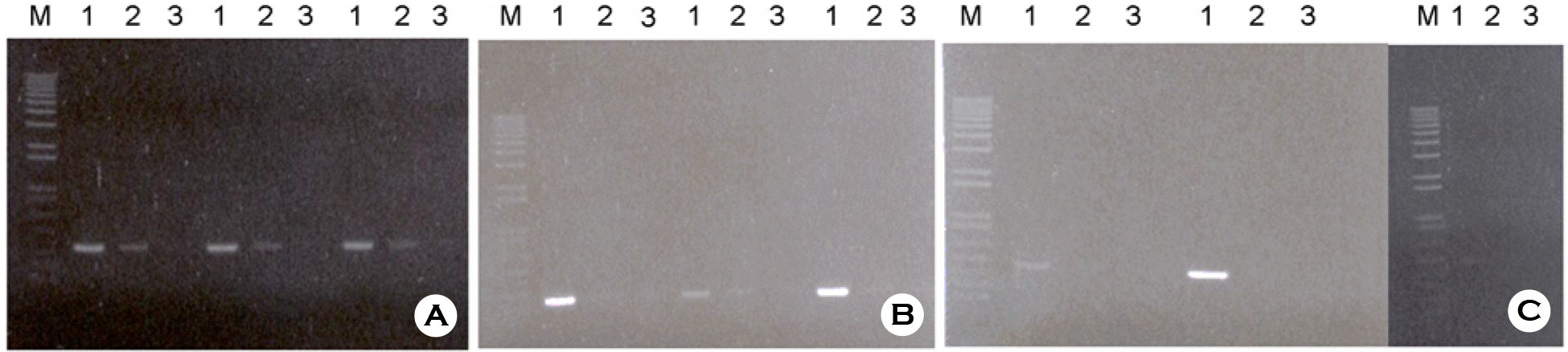
Figure 4.
Multiplex PCR Amplification with each different plasmid DNA template (1, HBV; 2, HCV; 3, HIV-1), with two combined templates (4, HBV + HCV; 5, HBV + HIV-1; 6, HCV + HIV-1), and with three different templates together (7, HBV + HCV + HIV-1).
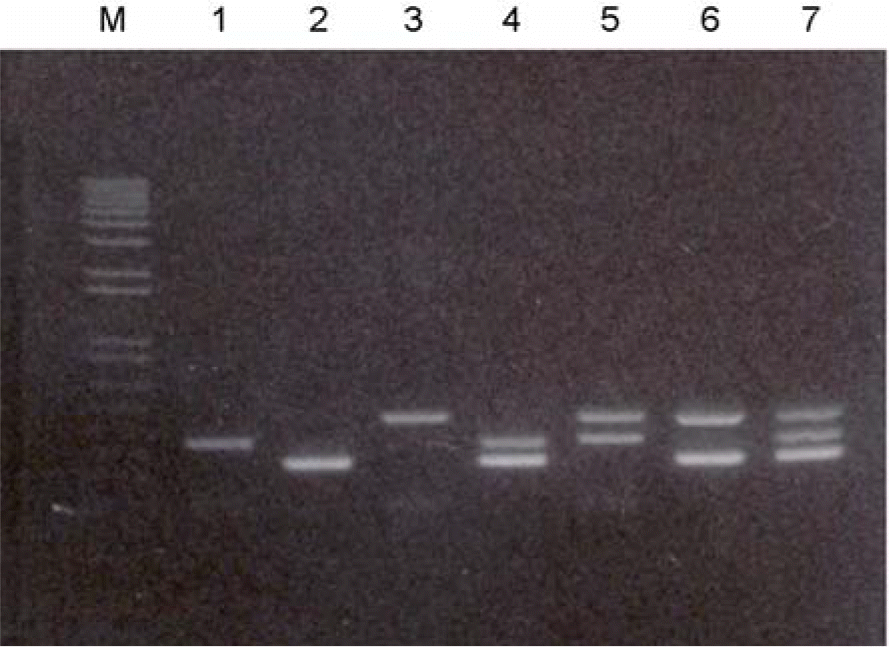
Figure 5.
Twenty different sera positively diagnosed by ELISA tests were amplified by multiplex PCR with 6 different primers: M, 100 bp marker; NC, negative control sera without any template DNA; C, three different plasmid DNAs such as HBV (A), HCV (B), HIV-1 (C), were amplified by same PCR for the control; each other lane is shown as each different sera samples amplified by multiplex PCR methods.
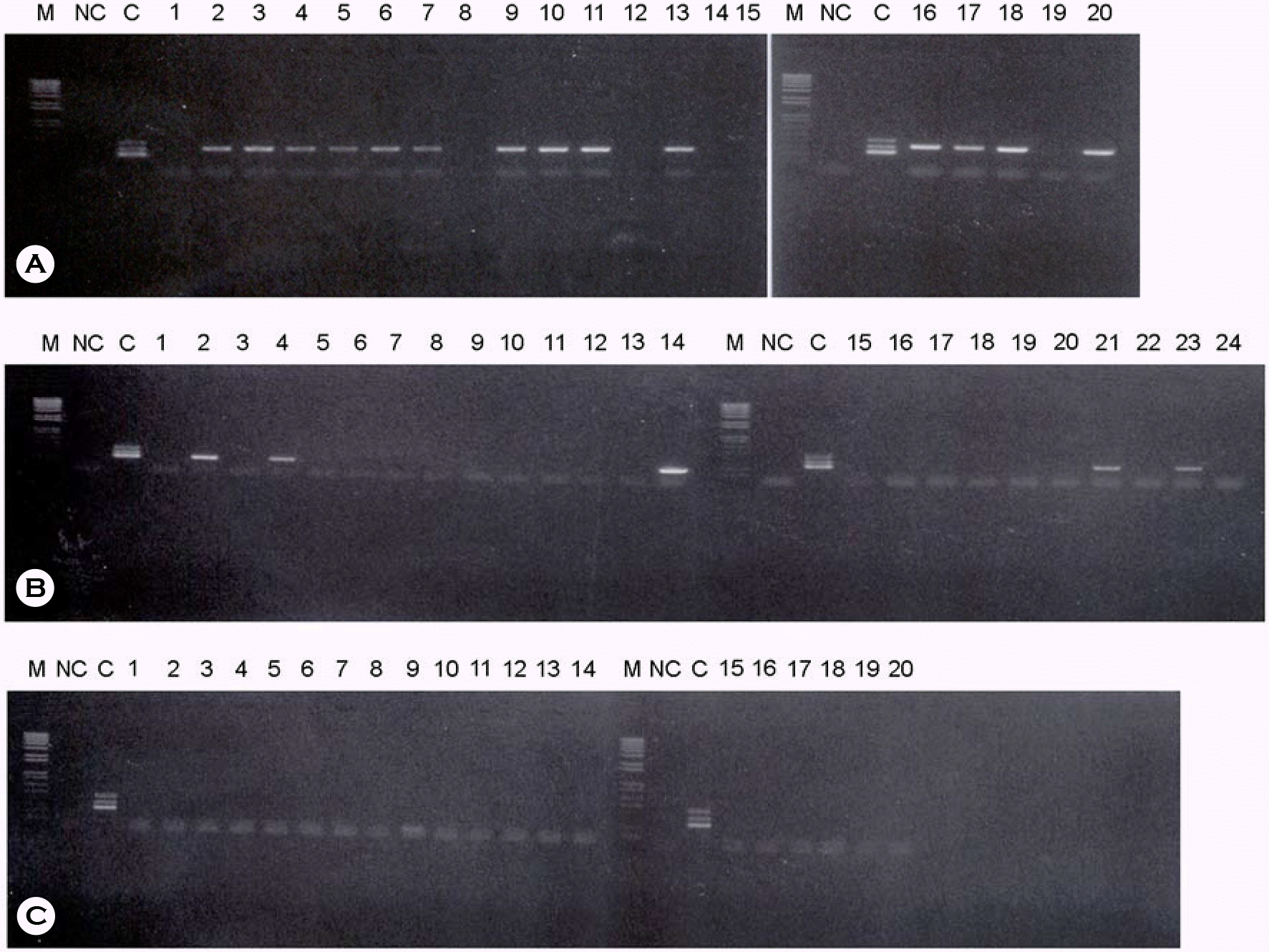
Figure 6.
Confirmation of the multiplex (RT)-PCR in one tube. Multiplex-PCR were performed with HBV-, HCV-positive sera, and sera containing HIV-1 plasmid DNA: Multiplex-PCR were performed with HBV-, HCV-positive sera, and HIV-1 plasmid: M, 100 bp size marker DNAs; lane 1, HBV-specific primer; lane 2, HCV-specific primer; lane 3, HIV-1 specific primer with HIV-1 plasmid DNA; lane 4, HBV and HCV specific primers; lane 5, HBV and HIV-1 specific primers; lane 6, HCV and HIV-1 specific primers; lanes 7 and 8, HBV, HCV, and HIV-1 specific primers were used, respectively. HBV sample number 10, HCV sample number 2 were used in the reaction. And also 1 μl of 1 μg/ml HIV-1 plasmid DNA was added into reaction
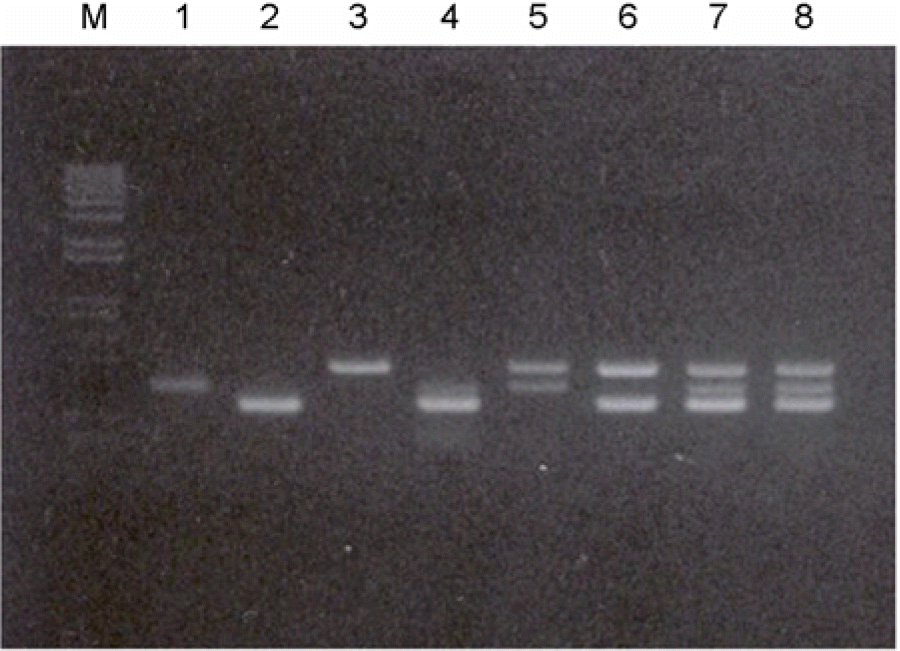
Table 1.
Oligonucleotides sequences used in the PCR and RT-PCR and products size after amplification
Table 2.
Specificity Tests by RT-PCR using viral-specific primers against non-infected human sera
Table 3.
Each sera identified as an infected with each virus were specifically screened by ELISA and (RT)-PCR
| HBV 결과 | HCV 결과 | HIV-1 결과 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ELISA∗ | PCR (그림 7) | ELISA∗∗ | PCR (그림 8) | ELISA∗∗∗ | PCR (그림 9) | |||||||
| HBV | HCV | HIV | HBV | HCV | HIV | HBV | HCV | HIV | ||||
| 음성대조액 1 | 0.056 | – | – | – | 0.026 | – | – | – | 0.043 | – | – | – |
| 음성대조액 2 | 0.045 | – | – | – | 0.022 | – | – | – | 0.041 | – | – | – |
| 양성대조액 1 | 2.568 | – | – | – | 2.441 | – | – | – | 2.776 | – | – | – |
| 양성대조액 2 | 2.527 | – | – | – | 2.409 | – | – | – | 2.934 | – | – | – |
| 검체 1 | 2.826 | – | – | – | 2.516 | – | – | – | 2.904 | – | – | – |
| 검체 2 | 2.800 | 양성 | – | – | 2.841 | – | 양성 | – | 2.861 | – | – | – |
| 검체 3 | 2.210 | 양성 | – | – | 2.794 | – | – | – | 2.759 | – | – | – |
| 검체 4 | 2.884 | 양성 | – | – | 2.746 | – | 양성 | – | 2.884 | – | – | – |
| 검체 5 | 0.475 | 양성 | – | – | 2.714 | – | – | – | 2.884 | – | – | – |
| 검체 6 | 2.842 | 양성 | – | – | 2.580 | – | – | – | 2.673 | – | – | – |
| 검체 7 | 2.735 | 양성 | – | – | 2.900 | – | – | – | 2.858 | – | – | – |
| 검체 8 | 2.331 | – | – | – | 2.848 | – | – | – | 2.936 | – | – | – |
| 검체 9 | 2.762 | 양성 | – | – | 2.912 | – | – | – | 2.553 | – | – | – |
| 검체 10 | 1.513 | 양성 | – | – | 2.926 | – | – | – | 2.727 | – | – | – |
| 검체 11 | 2.444 | 양성 | – | – | 2.762 | – | – | – | 2.887 | – | – | – |
| 검체 12 | 2.906 | – | – | – | 2.492 | – | – | – | 2.827 | – | – | – |
| 검체 13 | 2.903 | 양성 | – | – | 2.685 | – | – | – | 2.639 | – | – | – |
| 검체 14 | 2.055 | – | – | – | 2.433 | – | 양성 | – | 2.948 | – | – | – |
| 검체 15 | 0.876 | – | – | – | 2.778 | – | – | – | 2.830 | – | – | – |
| 검체 16 | 0.811 | 양성 | – | – | 1.321 | – | – | – | 2.738 | – | – | – |
| 검체 17 | 2.294 | 양성 | – | – | 2.773 | – | – | – | 2.973 | – | – | – |
| 검체 18 | 2.581 | 양성 | – | – | 2.939 | – | – | – | 2.909 | – | – | – |
| 검체 19 | 3.138 | – | – | – | 2.834 | – | – | – | 2.895 | – | – | – |
| 검체 20 | 2.572 | 양성 | – | – | 2.900 | – | – | – | 2.621 | – | – | – |
| 검체 21 | 0.054 | – | – | – | 2.407 | – | 양성 | – | 0.056 | – | – | – |
| 검체 22 | 0.054 | – | – | – | 2.931 | – | – | – | 0.070 | – | – | – |
| 검체 23 | 0.056 | – | – | – | 3.075 | – | 양성 | – | 0.042 | – | – | – |
| 검체 24 | 0.068 | – | – | – | 2.145 | – | – | – | 0.087 | – | – | – |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download