Abstract
Swine hepatitis E virus (HEV) has been reported as a new zoonotic agent due to its close genomic resemblance to the human HEV. Recently this virus is indicated as one of the important pathogens in xenotransplantation that uses pig as a donor animal. We carried out to investigate the prevalence of HEV infections among the pigs and human population in Chungnam region using a nested RT-PCR for detection of a part of HEV ORF2 gene. The sequences of the amplified DNA were analyzed and the genetical divergency were characterized. A total of 18 HEV strains, comprising 16 strains from pig and 2 strains from human, were genetically isolated from the fecal and serum samples. Among the isolates, 5 strains (2.5%) were detected from 200 swine sera and 2 strains (2.0%) from 100 human sera. All of the 16 swine strains were isolated from the pigs at 3 month of age, but none of age groups revealed the positive for swine HEV RNA. In comparison of the nucleotide sequence between 16 swine HEV and 2 human HEV isolates, the range of identities was 91.5% to 100%. Two human HEV isolates shared 99.7% homology. In phylogenetic analysis, all of the isolates were classified into genotype III, and the 18 isolates were also closely related to the prototype of swine HEV and human HEV strains isolated in the United States and others recently identified from swine in Japan and Netherland.
Go to : 
References
1). Aggarwal R, Krawczynski K. Hepatitis E: an overview and recent advances in clinical and laboratory research. J Gastroenterol Hepatol. 15:9–20. 2000.

2). Ahn JM, Kang SG, Lee DY, Shin SJ, Yoo HS. Identification of novel human hepatits (HEV) isolates and determination of the seroprevalence of HEV in Korea. J Clin Microbiol. 43(7):3042–3048. 2005.
3). Arankalle VA, Chobe LP, Joshi MV, Chadha MS, Biduth K, Walimbe AM. Human and swine hepatitis E viruses from Western India belong to different genotypes. J Hepato. 36:417–425. 2002.

4). Arankalle VA, Joshi MV, Kulkarni AM, Gandhe SS, Chobe LP, Rautmare SS, Mishra AC, Padbidri VS. Prevalence of anti-hepatitis E virus antibodies in different Indian animal species. J Viral Hepat. 8:223–227. 2001.

5). Berke T, Matson DO. Reclassification of the Caliciviridae into distinct genera and excluion of hepatits E virus from the family on the basis of comparative phylogenetic analysis. Arch Virol. 145:1421–1436. 2000.
6). Chandler JD, Riddell MA, Li F, Love RJ, Anderson DA. Serological evidence for swine hepatitis E virus infection in Australian pig herds. Vet Microbiol. 68:95–105. 1999.

7). Choi CS, Ha SK, Chae CH. Development of nested RT-PCR for the detection of swine hepatitis E virus in formalin-fixed, paraffin-embedded tissues and comparison with in situ hybridization. Journal of Virological Methods. 115:67–71. 2004.

8). Favorov MO, Kosoy MY, Tsarev SA, Childs JE, Margolis HS. Prevalence of antibody to hepatitis E virus among rodents in the United States. J Infect Dis. 181:449–455. 2000.

9). Halbur PG, Kasorndorkbua C, Gilbert C, Guenette D, Potters MB, Purcell RH, Emerson SU, Toth TE, Meng XJ. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J Clin Microbiol. 39:918–923. 2001.

10). Hsieh SY, Meng XJ, Wu YH, Liu ST, Tam AW, Lin DY, Liaw YF. Identity of a novel swine hepatitis E virus in Taiwan forming a monophyletic group with Taiwan isolates of human hepatitis E virus. J Clin Microbiol. 37:3828–3834. 1999.

11). Huang FF, Haqshenas G, Guenette DK, Halbur PG, Schommer SK, Pierson FW, Toth TE, Meng XJ. Detection by reverse transcription-PCR and genetic characterization of field isolates of swine hepatitis E virus from pigs in different geographic regions of the United States. J Clin Mincrobiol. 40:1326–1332. 2002.

12). Meng XJ. Swine hepatitis E virus: cross-species infection and risk in xenotransplantation. Curr Top Crobiol Immunol. 278:185–216. 2003.
13). Meng XJ, Dea S, Engle RE, Friendship R, Lyoo YS, Sirinarumitr T, Urairong K, Wang D, Yoo D, Zhang Y, Purcell RH, Emerson SU. Prevalence of antibodies to the hepatitis E virus in pigs from countries where hepatitis E is common or rare in the human population. J Med Virol. 59:297–302. 1999.
14). Meng XJ, Halbur PG, Haynes JS, Tsareva TS, Bruna JD, Royer RL, Purcell RH, Emerson SU. Experimental infection of pigs with the newly identified swine hepatitis E virus (swine HEV), but not with human strains of HEV. Arch. Virol. 143:1405–1415. 1998.

15). Meng XJ, Halbur PG, Shapiro MS, Govindarajan S, Bruna JD, Musgahwar IK, Purcell RH, Emerson SU. Genetic and experimental evidence for cross-species infection by the swine hepatitis E virus. J Virol. 72:9714–9721. 1998.
16). Meng XJ, Purcell RH, Halbur PG, Lehman JR, Webb DM, Tsareva TS, Haynes JS, Thacker BJ, Emerson SU. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci USA. 94:9860–9865. 1997.

17). Meng XJ, Wiseman B, Elvinger F, Guenette DK, Toth TE, Engle RE, Emerson SU, Purcell RH. Prevalence of antibodies to hepatitis E virus in veterinarians working with swine and in normal blood donors in the United States and other countries. J Clin Microbiol. 40:117–122. 2002.

18). Okamoto H, Takahashi M, Nishizawa T, Fukai K, Muramatsu U, Yoshikawa A. Analysis of the complete genome of indigenous swine hepatitis E virus isolated in Japan. Biochem. Biophys. Res Commun. 289:929–936. 2001.

19). Poel VD, Verschoor F, Heide RVD, Herrera MI, Vivo A, Kooreman M, Husman DA. Hepatitis E virus sequences in swine related to sequences in humans, the Netherlands. Emerg Infect Dis. 7:970–976. 2001.
20). Pringle CR. Virus taxonomy-San Diego 1998. Arch Virol. 143:1449–1459. 1998.
21). Purcell RH. Hepatitis E virus. p. 2831–2843. In. Fields BN, Knipe DM, Howley PM, editors. (ed.),. Fields virology. 4rd ed.vol.2:Lippincott-Raven Publishers;Philadelphia: 2001.
22). Reyes GR. Overview of the epidemiology and biology of the hepatitis E virus. p. 239–258. In. Willsen RA, editor. (ed.),. Viral hepatitis. Marcel Dekker, Inc.;New York, N.Y.: 1997.
23). Schlauder GG, Dawson GJ, Erker JC, Kwo PY, Knigge MF, Smally DL, Rosenblatt JE, Desai SM, Mushahwar IK. The sequence and phylogenetic analysis of a novel hepatitis E virus isolated from a patient with acute hepatitis reported in the United States. J Gen Virol. 79:447–456. 1998.

24). Takahashi K, Kang JH, Ohnishi S, Hino K, Miyakawa H, Miyakawa Y, Maekubo H, Mishiro S. Full-length sequences of six hepatitis E virus isolates of genotypes III and IV from patients with sporadic acute or fulminant hepatitis in Japan. Intervirology. 46(5):308–314. 2003.

25). Takahashi M, Nishizawa T, Miyajima H, Gotanda Y, Iita T, Tsuda F, Okamoto H. Swine hepatitis E virus strains in Japan form four phylogenetic clusters comparable with those of Japanese isolates of human hepatitis E virus. J Gen Virol. 84:851–862. 2003.

26). Wang Y, Zhang H, Li Z, Gu W, Lan H, Hao W, Ling R, Li H, Harrison TJ. Detection of sporadic cases of hepatitis E virus (HEV) infection in China using immunoassays based on recombinant open reading frame 2 and 3 polypeptides from HEV genotype 4. J Clin Microbiol. 39:4370–4379. 2001.

27). Wang Y, Zhang H, Xia N, et al. Prevalence, isolation, and partial sequence analysis of hepatitis E virus from domestic animals in China. J Med Virol. 67:516–521. 2002.

29). Wu JC, Chen CM, Chiang TY, Tsai WH, Jeng WJ, Sheen IJ, Kin CC, Meng XJ. Spread of hepatitis E virus among different-aged pigs: two-year survey in Taiwan. J Med Virol. 66:488–492. 2002.

30). Yoo D, Giulivi A. Xenotransplantation and the potential risk of xenogeneic transmission of porcine viruses. Can J Vet Res. 64:193–203. 2000.
Go to : 
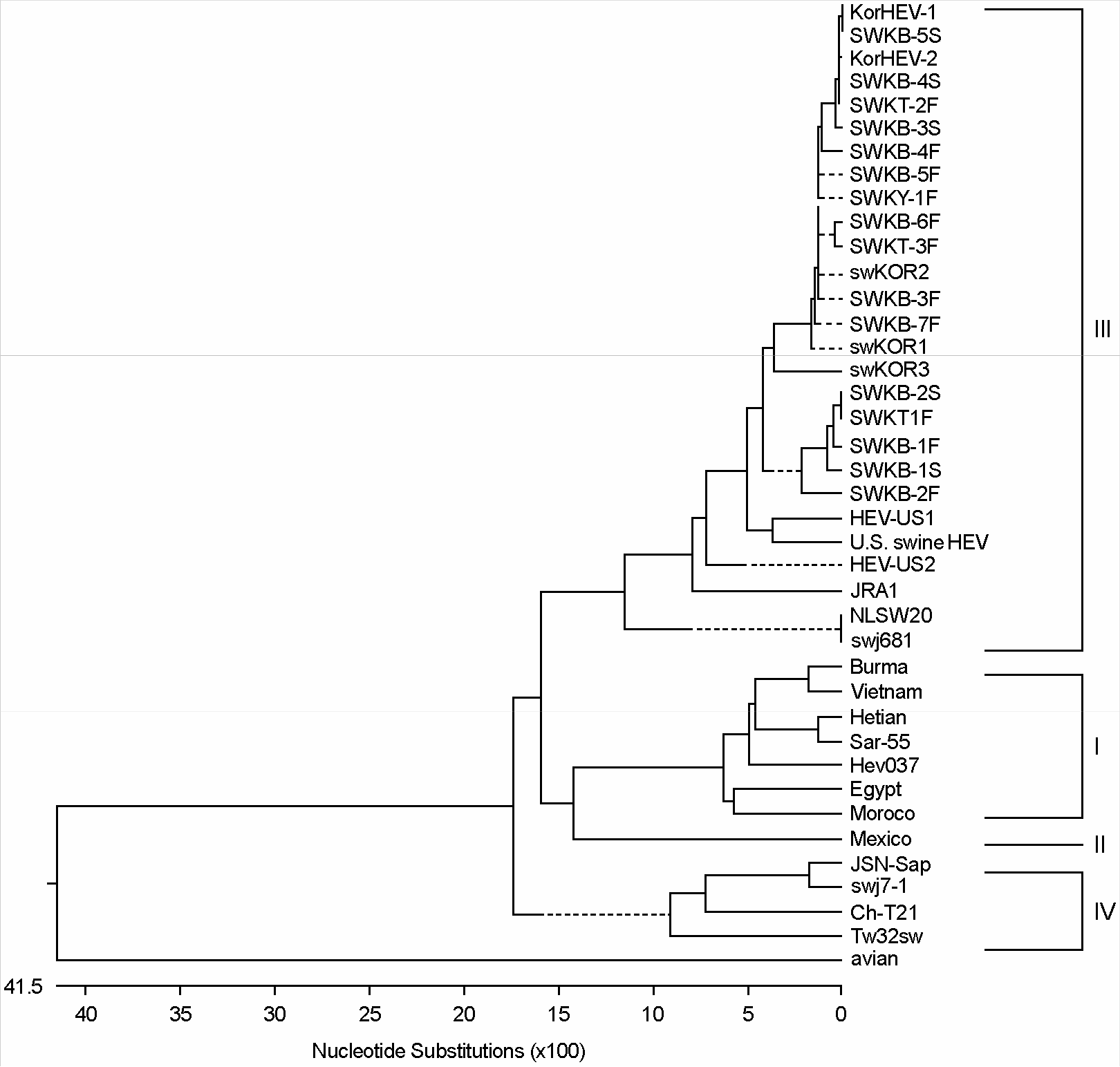 | Figure 1.Phylogenetic tree analysis of Korean isolates of swine HEV by comparison of the conserved 323 bp (6020∼6342 nt) of ORF2 in the various swine and human HEV isolates. Groups; I, II, III and IV. |
Table 1.
Detection of HEV RNA in the specimens from the pigs and humans
| Species | Age groups | No. of samples tested | No. of positive samples (%) | Designation of isolates | |
|---|---|---|---|---|---|
| Feces | Serum | ||||
| Pig |
2a 3 4 5 >7 (sows) Subtotal |
29 67 49 25 30 200 |
0 11 (16.4) 0 0 0 11 (5.5) |
0 5 (7.5) 0 0 0 5 (2.5) |
SWKB1F, SWKB2F, SWKB3F, SWKB4F, SWKB5F, SWKB6F, SWKB7F, SWKB1S, SWKB2S, SWKB3S, SWKB4S, SWKB5S SWKT1F, SWKT2F, SWKT3F, SWKY1F |
| Human | 30∼50b | 100 | NT | 2 (2.0) | KorHEV-1, KorHEV-2 |
Table 2.
Pairwise comparison of the nucleotide identity of partial ORF2 gene (6020∼6342 nt) of 18 HEV isolates identified in this study and other selected strains of HEV worldwidea
a References for other HEV strains used in comparisons: Hev037 (X98292, India), Sar-55 (M80581, Pakistan), Vietnam (AF170450), Burma (M73218), JRA1 (AP003430, Japan), Hetian (L08816, China), Ch-T21 (AF151963, China), Tw32sw (AF117280, Taiwan), Morocco (AF065061), Mexico (M74506), U.S. swine HEV (AF082843), HEV-US1 (AF060668, U.S.), HEV-US2 (AF060669, U.S.), avian (AY043166, U.S.). swj7–1 (AB094219, Japan), JSN-Sap (AB091395, Japan), swj681 (AB073910, Japan), NLSW20 (AF336290, Netherlands), Egypt (AF051351), swKOR1 (AF516178, Korea), swKOR2 (AF516179, Korea), and swKOR3 (AF527942, Korea)




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download